Sirui Ding
Rethinking the Understanding Ability across LLMs through Mutual Information
May 25, 2025Abstract:Recent advances in large language models (LLMs) have revolutionized natural language processing, yet evaluating their intrinsic linguistic understanding remains challenging. Moving beyond specialized evaluation tasks, we propose an information-theoretic framework grounded in mutual information (MI) to achieve this. We formalize the understanding as MI between an input sentence and its latent representation (sentence-level MI), measuring how effectively input information is preserved in latent representation. Given that LLMs learn embeddings for individual tokens, we decompose sentence-level MI into token-level MI between tokens and sentence embeddings, establishing theoretical bounds connecting these measures. Based on this foundation, we theoretically derive a computable lower bound for token-level MI using Fano's inequality, which directly relates to token-level recoverability-the ability to predict original tokens from sentence embedding. We implement this recoverability task to comparatively measure MI across different LLMs, revealing that encoder-only models consistently maintain higher information fidelity than their decoder-only counterparts, with the latter exhibiting a distinctive late-layer "forgetting" pattern where mutual information is first enhanced and then discarded. Moreover, fine-tuning to maximize token-level recoverability consistently improves understanding ability of LLMs on tasks without task-specific supervision, demonstrating that mutual information can serve as a foundation for understanding and improving language model capabilities.
GRAPHGPT-O: Synergistic Multimodal Comprehension and Generation on Graphs
Feb 17, 2025



Abstract:The rapid development of Multimodal Large Language Models (MLLMs) has enabled the integration of multiple modalities, including texts and images, within the large language model (LLM) framework. However, texts and images are usually interconnected, forming a multimodal attributed graph (MMAG). It is underexplored how MLLMs can incorporate the relational information (\textit{i.e.}, graph structure) and semantic information (\textit{i.e.,} texts and images) on such graphs for multimodal comprehension and generation. In this paper, we propose GraphGPT-o, which supports omni-multimodal understanding and creation on MMAGs. We first comprehensively study linearization variants to transform semantic and structural information as input for MLLMs. Then, we propose a hierarchical aligner that enables deep graph encoding, bridging the gap between MMAGs and MLLMs. Finally, we explore the inference choices, adapting MLLM to interleaved text and image generation in graph scenarios. Extensive experiments on three datasets from different domains demonstrate the effectiveness of our proposed method. Datasets and codes will be open-sourced upon acceptance.
Large Language Models for Disease Diagnosis: A Scoping Review
Aug 27, 2024Abstract:Automatic disease diagnosis has become increasingly valuable in clinical practice. The advent of large language models (LLMs) has catalyzed a paradigm shift in artificial intelligence, with growing evidence supporting the efficacy of LLMs in diagnostic tasks. Despite the growing attention in this field, many critical research questions remain under-explored. For instance, what diseases and LLM techniques have been investigated for diagnostic tasks? How can suitable LLM techniques and evaluation methods be selected for clinical decision-making? To answer these questions, we performed a comprehensive analysis of LLM-based methods for disease diagnosis. This scoping review examined the types of diseases, associated organ systems, relevant clinical data, LLM techniques, and evaluation methods reported in existing studies. Furthermore, we offered guidelines for data preprocessing and the selection of appropriate LLM techniques and evaluation strategies for diagnostic tasks. We also assessed the limitations of current research and delineated the challenges and future directions in this research field. In summary, our review outlined a blueprint for LLM-based disease diagnosis, helping to streamline and guide future research endeavors.
Interpretable Differential Diagnosis with Dual-Inference Large Language Models
Jul 10, 2024
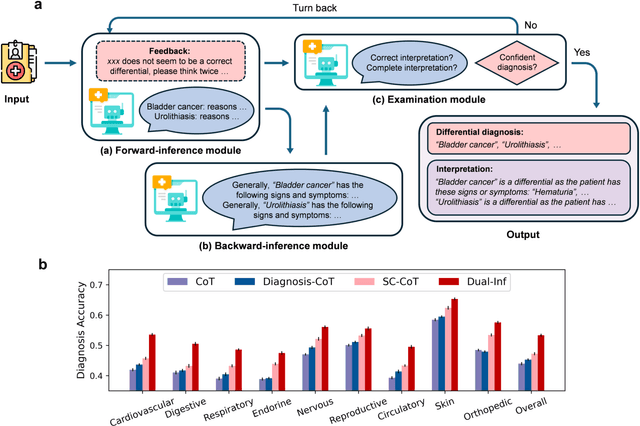


Abstract:Methodological advancements to automate the generation of differential diagnosis (DDx) to predict a list of potential diseases as differentials given patients' symptom descriptions are critical to clinical reasoning and applications such as decision support. However, providing reasoning or interpretation for these differential diagnoses is more meaningful. Fortunately, large language models (LLMs) possess powerful language processing abilities and have been proven effective in various related tasks. Motivated by this potential, we investigate the use of LLMs for interpretable DDx. First, we develop a new DDx dataset with expert-derived interpretation on 570 public clinical notes. Second, we propose a novel framework, named Dual-Inf, that enables LLMs to conduct bidirectional inference for interpretation. Both human and automated evaluation demonstrate the effectiveness of Dual-Inf in predicting differentials and diagnosis explanations. Specifically, the performance improvement of Dual-Inf over the baseline methods exceeds 32% w.r.t. BERTScore in DDx interpretation. Furthermore, experiments verify that Dual-Inf (1) makes fewer errors in interpretation, (2) has great generalizability, (3) is promising for rare disease diagnosis and explanation.
Understanding Different Design Choices in Training Large Time Series Models
Jun 20, 2024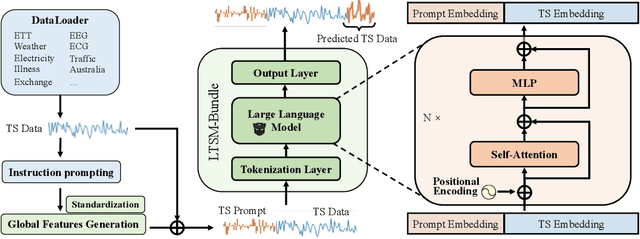
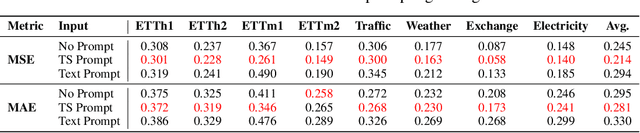
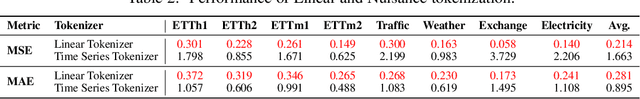
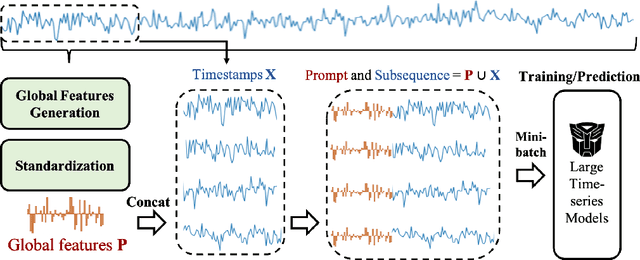
Abstract:Inspired by Large Language Models (LLMs), Time Series Forecasting (TSF), a long-standing task in time series analysis, is undergoing a transition towards Large Time Series Models (LTSMs), aiming to train universal transformer-based models for TSF. However, training LTSMs on heterogeneous time series data poses unique challenges, including diverse frequencies, dimensions, and patterns across datasets. Recent endeavors have studied and evaluated various design choices aimed at enhancing LTSM training and generalization capabilities, spanning pre-processing techniques, model configurations, and dataset configurations. In this work, we comprehensively analyze these design choices and aim to identify the best practices for training LTSM. Moreover, we propose \emph{time series prompt}, a novel statistical prompting strategy tailored to time series data. Furthermore, based on the observations in our analysis, we introduce \texttt{LTSM-bundle}, which bundles the best design choices we have identified. Empirical results demonstrate that \texttt{LTSM-bundle} achieves superior zero-shot and few-shot performances compared to state-of-the-art LSTMs and traditional TSF methods on benchmark datasets.
MolecularGPT: Open Large Language Model (LLM) for Few-Shot Molecular Property Prediction
Jun 18, 2024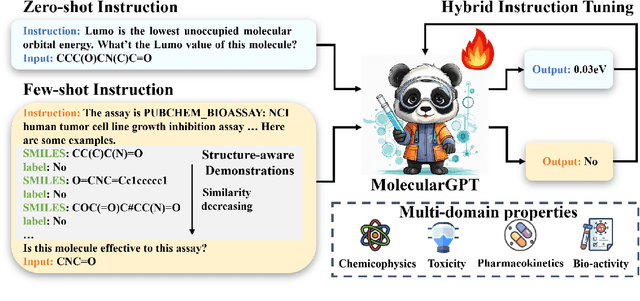
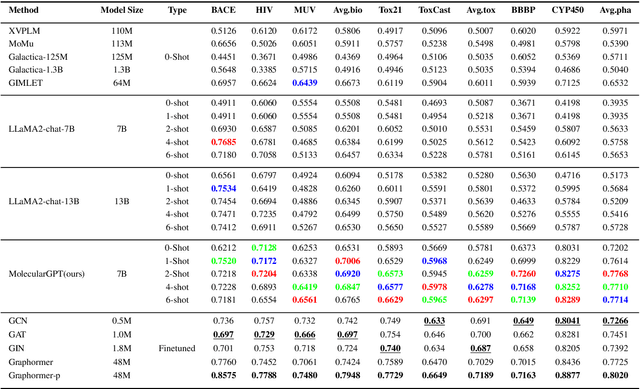
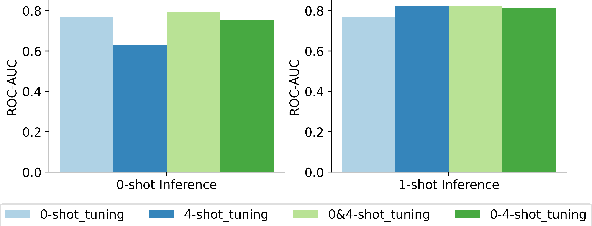

Abstract:Molecular property prediction (MPP) is a fundamental and crucial task in drug discovery. However, prior methods are limited by the requirement for a large number of labeled molecules and their restricted ability to generalize for unseen and new tasks, both of which are essential for real-world applications. To address these challenges, we present MolecularGPT for few-shot MPP. From a perspective on instruction tuning, we fine-tune large language models (LLMs) based on curated molecular instructions spanning over 1000 property prediction tasks. This enables building a versatile and specialized LLM that can be adapted to novel MPP tasks without any fine-tuning through zero- and few-shot in-context learning (ICL). MolecularGPT exhibits competitive in-context reasoning capabilities across 10 downstream evaluation datasets, setting new benchmarks for few-shot molecular prediction tasks. More importantly, with just two-shot examples, MolecularGPT can outperform standard supervised graph neural network methods on 4 out of 7 datasets. It also excels state-of-the-art LLM baselines by up to 16.6% increase on classification accuracy and decrease of 199.17 on regression metrics (e.g., RMSE) under zero-shot. This study demonstrates the potential of LLMs as effective few-shot molecular property predictors. The code is available at https://github.com/NYUSHCS/MolecularGPT.
UniGLM: Training One Unified Language Model for Text-Attributed Graphs
Jun 17, 2024



Abstract:Representation learning on text-attributed graphs (TAGs), where nodes are represented by textual descriptions, is crucial for textual and relational knowledge systems and recommendation systems. Currently, state-of-the-art embedding methods for TAGs primarily focus on fine-tuning language models (e.g., BERT) using structure-aware training signals. While effective, these methods are tailored for individual TAG and cannot generalize across various graph scenarios. Given the shared textual space, leveraging multiple TAGs for joint fine-tuning, aligning text and graph structure from different aspects, would be more beneficial. Motivated by this, we introduce a novel Unified Graph Language Model (UniGLM) framework, the first graph embedding model that generalizes well to both in-domain and cross-domain TAGs. Specifically, UniGLM is trained over multiple TAGs with different domains and scales using self-supervised contrastive learning. UniGLM includes an adaptive positive sample selection technique for identifying structurally similar nodes and a lazy contrastive module that is devised to accelerate training by minimizing repetitive encoding calculations. Extensive empirical results across 9 benchmark TAGs demonstrate UniGLM's efficacy against leading embedding baselines in terms of generalization (various downstream tasks and backbones) and transfer learning (in and out of domain scenarios). The code is available at https://github.com/NYUSHCS/UniGLM.
Multi-Task Learning for Post-transplant Cause of Death Analysis: A Case Study on Liver Transplant
Mar 30, 2023Abstract:Organ transplant is the essential treatment method for some end-stage diseases, such as liver failure. Analyzing the post-transplant cause of death (CoD) after organ transplant provides a powerful tool for clinical decision making, including personalized treatment and organ allocation. However, traditional methods like Model for End-stage Liver Disease (MELD) score and conventional machine learning (ML) methods are limited in CoD analysis due to two major data and model-related challenges. To address this, we propose a novel framework called CoD-MTL leveraging multi-task learning to model the semantic relationships between various CoD prediction tasks jointly. Specifically, we develop a novel tree distillation strategy for multi-task learning, which combines the strength of both the tree model and multi-task learning. Experimental results are presented to show the precise and reliable CoD predictions of our framework. A case study is conducted to demonstrate the clinical importance of our method in the liver transplant.
Towards Fair Patient-Trial Matching via Patient-Criterion Level Fairness Constraint
Mar 24, 2023



Abstract:Clinical trials are indispensable in developing new treatments, but they face obstacles in patient recruitment and retention, hindering the enrollment of necessary participants. To tackle these challenges, deep learning frameworks have been created to match patients to trials. These frameworks calculate the similarity between patients and clinical trial eligibility criteria, considering the discrepancy between inclusion and exclusion criteria. Recent studies have shown that these frameworks outperform earlier approaches. However, deep learning models may raise fairness issues in patient-trial matching when certain sensitive groups of individuals are underrepresented in clinical trials, leading to incomplete or inaccurate data and potential harm. To tackle the issue of fairness, this work proposes a fair patient-trial matching framework by generating a patient-criterion level fairness constraint. The proposed framework considers the inconsistency between the embedding of inclusion and exclusion criteria among patients of different sensitive groups. The experimental results on real-world patient-trial and patient-criterion matching tasks demonstrate that the proposed framework can successfully alleviate the predictions that tend to be biased.
PheME: A deep ensemble framework for improving phenotype prediction from multi-modal data
Mar 19, 2023Abstract:Detailed phenotype information is fundamental to accurate diagnosis and risk estimation of diseases. As a rich source of phenotype information, electronic health records (EHRs) promise to empower diagnostic variant interpretation. However, how to accurately and efficiently extract phenotypes from the heterogeneous EHR data remains a challenge. In this work, we present PheME, an Ensemble framework using Multi-modality data of structured EHRs and unstructured clinical notes for accurate Phenotype prediction. Firstly, we employ multiple deep neural networks to learn reliable representations from the sparse structured EHR data and redundant clinical notes. A multi-modal model then aligns multi-modal features onto the same latent space to predict phenotypes. Secondly, we leverage ensemble learning to combine outputs from single-modal models and multi-modal models to improve phenotype predictions. We choose seven diseases to evaluate the phenotyping performance of the proposed framework. Experimental results show that using multi-modal data significantly improves phenotype prediction in all diseases, the proposed ensemble learning framework can further boost the performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge