MolecularGPT: Open Large Language Model (LLM) for Few-Shot Molecular Property Prediction
Paper and Code
Jun 18, 2024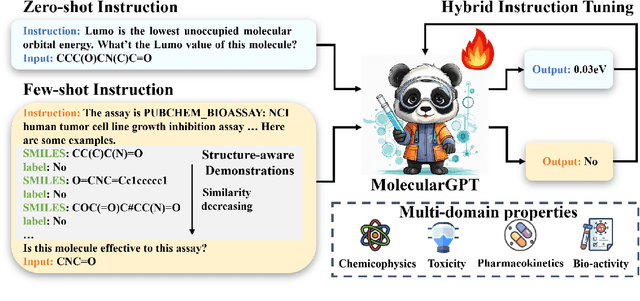
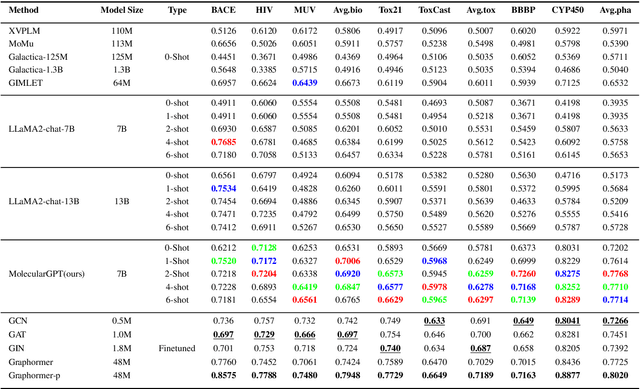
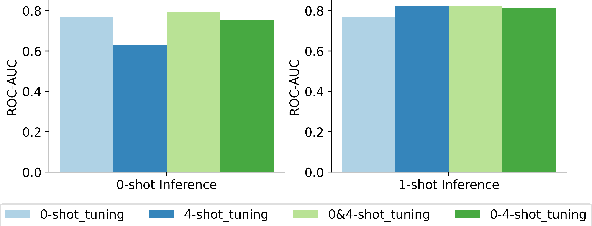

Molecular property prediction (MPP) is a fundamental and crucial task in drug discovery. However, prior methods are limited by the requirement for a large number of labeled molecules and their restricted ability to generalize for unseen and new tasks, both of which are essential for real-world applications. To address these challenges, we present MolecularGPT for few-shot MPP. From a perspective on instruction tuning, we fine-tune large language models (LLMs) based on curated molecular instructions spanning over 1000 property prediction tasks. This enables building a versatile and specialized LLM that can be adapted to novel MPP tasks without any fine-tuning through zero- and few-shot in-context learning (ICL). MolecularGPT exhibits competitive in-context reasoning capabilities across 10 downstream evaluation datasets, setting new benchmarks for few-shot molecular prediction tasks. More importantly, with just two-shot examples, MolecularGPT can outperform standard supervised graph neural network methods on 4 out of 7 datasets. It also excels state-of-the-art LLM baselines by up to 16.6% increase on classification accuracy and decrease of 199.17 on regression metrics (e.g., RMSE) under zero-shot. This study demonstrates the potential of LLMs as effective few-shot molecular property predictors. The code is available at https://github.com/NYUSHCS/MolecularGPT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge