Yuyan Liu
What Roles can Spatial Modulation and Space Shift Keying Play in LEO Satellite-Assisted Communication?
Sep 26, 2024



Abstract:In recent years, the rapid evolution of satellite communications play a pivotal role in addressing the ever-increasing demand for global connectivity, among which the Low Earth Orbit (LEO) satellites attract a great amount of attention due to their low latency and high data throughput capabilities. Based on this, we explore spatial modulation (SM) and space shift keying (SSK) designs as pivotal techniques to enhance spectral efficiency (SE) and bit-error rate (BER) performance in the LEO satellite-assisted multiple-input multiple-output (MIMO) systems. The various performance analysis of these designs are presented in this paper, revealing insightful findings and conclusions through analytical methods and Monte Carlo simulations with perfect and imperfect channel state information (CSI) estimation. The results provide a comprehensive analysis of the merits and trade-offs associated with the investigated schemes, particularly in terms of BER, computational complexity, and SE. This analysis underscores the potential of both schemes as viable candidates for future 6G LEO satellite-assisted wireless communication systems.
MolecularGPT: Open Large Language Model (LLM) for Few-Shot Molecular Property Prediction
Jun 18, 2024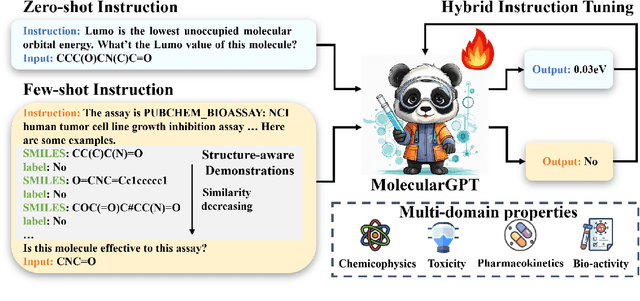
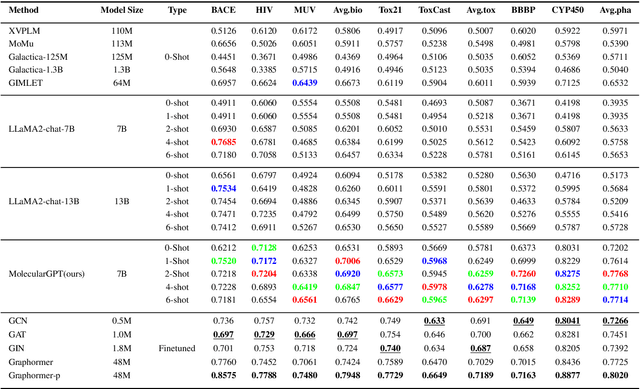
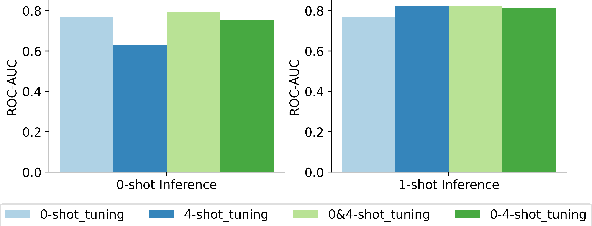

Abstract:Molecular property prediction (MPP) is a fundamental and crucial task in drug discovery. However, prior methods are limited by the requirement for a large number of labeled molecules and their restricted ability to generalize for unseen and new tasks, both of which are essential for real-world applications. To address these challenges, we present MolecularGPT for few-shot MPP. From a perspective on instruction tuning, we fine-tune large language models (LLMs) based on curated molecular instructions spanning over 1000 property prediction tasks. This enables building a versatile and specialized LLM that can be adapted to novel MPP tasks without any fine-tuning through zero- and few-shot in-context learning (ICL). MolecularGPT exhibits competitive in-context reasoning capabilities across 10 downstream evaluation datasets, setting new benchmarks for few-shot molecular prediction tasks. More importantly, with just two-shot examples, MolecularGPT can outperform standard supervised graph neural network methods on 4 out of 7 datasets. It also excels state-of-the-art LLM baselines by up to 16.6% increase on classification accuracy and decrease of 199.17 on regression metrics (e.g., RMSE) under zero-shot. This study demonstrates the potential of LLMs as effective few-shot molecular property predictors. The code is available at https://github.com/NYUSHCS/MolecularGPT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge