Xia Hu
RAPO: Risk-Aware Preference Optimization for Generalizable Safe Reasoning
Feb 04, 2026Abstract:Large Reasoning Models (LRMs) have achieved tremendous success with their chain-of-thought (CoT) reasoning, yet also face safety issues similar to those of basic language models. In particular, while algorithms are designed to guide them to deliberately refuse harmful prompts with safe reasoning, this process often fails to generalize against diverse and complex jailbreak attacks. In this work, we attribute these failures to the generalization of the safe reasoning process, particularly their insufficiency against complex attack prompts. We provide both theoretical and empirical evidence to show the necessity of a more sufficient safe reasoning process to defend against advanced attack prompts. Building on this insight, we propose a Risk-Aware Preference Optimization (RAPO) framework that enables LRM to adaptively identify and address the safety risks with appropriate granularity in its thinking content. Extensive experiments demonstrate that RAPO successfully generalizes multiple LRMs' safe reasoning adaptively across diverse attack prompts whilst preserving general utility, contributing a robust alignment technique for LRM safety. Our code is available at https://github.com/weizeming/RAPO.
LPS-Bench: Benchmarking Safety Awareness of Computer-Use Agents in Long-Horizon Planning under Benign and Adversarial Scenarios
Feb 03, 2026Abstract:Computer-use agents (CUAs) that interact with real computer systems can perform automated tasks but face critical safety risks. Ambiguous instructions may trigger harmful actions, and adversarial users can manipulate tool execution to achieve malicious goals. Existing benchmarks mostly focus on short-horizon or GUI-based tasks, evaluating on execution-time errors but overlooking the ability to anticipate planning-time risks. To fill this gap, we present LPS-Bench, a benchmark that evaluates the planning-time safety awareness of MCP-based CUAs under long-horizon tasks, covering both benign and adversarial interactions across 65 scenarios of 7 task domains and 9 risk types. We introduce a multi-agent automated pipeline for scalable data generation and adopt an LLM-as-a-judge evaluation protocol to assess safety awareness through the planning trajectory. Experiments reveal substantial deficiencies in existing CUAs' ability to maintain safe behavior. We further analyze the risks and propose mitigation strategies to improve long-horizon planning safety in MCP-based CUA systems. We open-source our code at https://github.com/tychenn/LPS-Bench.
Interpreting Emergent Extreme Events in Multi-Agent Systems
Jan 28, 2026Abstract:Large language model-powered multi-agent systems have emerged as powerful tools for simulating complex human-like systems. The interactions within these systems often lead to extreme events whose origins remain obscured by the black box of emergence. Interpreting these events is critical for system safety. This paper proposes the first framework for explaining emergent extreme events in multi-agent systems, aiming to answer three fundamental questions: When does the event originate? Who drives it? And what behaviors contribute to it? Specifically, we adapt the Shapley value to faithfully attribute the occurrence of extreme events to each action taken by agents at different time steps, i.e., assigning an attribution score to the action to measure its influence on the event. We then aggregate the attribution scores along the dimensions of time, agent, and behavior to quantify the risk contribution of each dimension. Finally, we design a set of metrics based on these contribution scores to characterize the features of extreme events. Experiments across diverse multi-agent system scenarios (economic, financial, and social) demonstrate the effectiveness of our framework and provide general insights into the emergence of extreme phenomena.
AgentDoG: A Diagnostic Guardrail Framework for AI Agent Safety and Security
Jan 26, 2026Abstract:The rise of AI agents introduces complex safety and security challenges arising from autonomous tool use and environmental interactions. Current guardrail models lack agentic risk awareness and transparency in risk diagnosis. To introduce an agentic guardrail that covers complex and numerous risky behaviors, we first propose a unified three-dimensional taxonomy that orthogonally categorizes agentic risks by their source (where), failure mode (how), and consequence (what). Guided by this structured and hierarchical taxonomy, we introduce a new fine-grained agentic safety benchmark (ATBench) and a Diagnostic Guardrail framework for agent safety and security (AgentDoG). AgentDoG provides fine-grained and contextual monitoring across agent trajectories. More Crucially, AgentDoG can diagnose the root causes of unsafe actions and seemingly safe but unreasonable actions, offering provenance and transparency beyond binary labels to facilitate effective agent alignment. AgentDoG variants are available in three sizes (4B, 7B, and 8B parameters) across Qwen and Llama model families. Extensive experimental results demonstrate that AgentDoG achieves state-of-the-art performance in agentic safety moderation in diverse and complex interactive scenarios. All models and datasets are openly released.
The Why Behind the Action: Unveiling Internal Drivers via Agentic Attribution
Jan 21, 2026Abstract:Large Language Model (LLM)-based agents are widely used in real-world applications such as customer service, web navigation, and software engineering. As these systems become more autonomous and are deployed at scale, understanding why an agent takes a particular action becomes increasingly important for accountability and governance. However, existing research predominantly focuses on \textit{failure attribution} to localize explicit errors in unsuccessful trajectories, which is insufficient for explaining the reasoning behind agent behaviors. To bridge this gap, we propose a novel framework for \textbf{general agentic attribution}, designed to identify the internal factors driving agent actions regardless of the task outcome. Our framework operates hierarchically to manage the complexity of agent interactions. Specifically, at the \textit{component level}, we employ temporal likelihood dynamics to identify critical interaction steps; then at the \textit{sentence level}, we refine this localization using perturbation-based analysis to isolate the specific textual evidence. We validate our framework across a diverse suite of agentic scenarios, including standard tool use and subtle reliability risks like memory-induced bias. Experimental results demonstrate that the proposed framework reliably pinpoints pivotal historical events and sentences behind the agent behavior, offering a critical step toward safer and more accountable agentic systems.
OpenRT: An Open-Source Red Teaming Framework for Multimodal LLMs
Jan 04, 2026Abstract:The rapid integration of Multimodal Large Language Models (MLLMs) into critical applications is increasingly hindered by persistent safety vulnerabilities. However, existing red-teaming benchmarks are often fragmented, limited to single-turn text interactions, and lack the scalability required for systematic evaluation. To address this, we introduce OpenRT, a unified, modular, and high-throughput red-teaming framework designed for comprehensive MLLM safety evaluation. At its core, OpenRT architects a paradigm shift in automated red-teaming by introducing an adversarial kernel that enables modular separation across five critical dimensions: model integration, dataset management, attack strategies, judging methods, and evaluation metrics. By standardizing attack interfaces, it decouples adversarial logic from a high-throughput asynchronous runtime, enabling systematic scaling across diverse models. Our framework integrates 37 diverse attack methodologies, spanning white-box gradients, multi-modal perturbations, and sophisticated multi-agent evolutionary strategies. Through an extensive empirical study on 20 advanced models (including GPT-5.2, Claude 4.5, and Gemini 3 Pro), we expose critical safety gaps: even frontier models fail to generalize across attack paradigms, with leading models exhibiting average Attack Success Rates as high as 49.14%. Notably, our findings reveal that reasoning models do not inherently possess superior robustness against complex, multi-turn jailbreaks. By open-sourcing OpenRT, we provide a sustainable, extensible, and continuously maintained infrastructure that accelerates the development and standardization of AI safety.
BLASST: Dynamic BLocked Attention Sparsity via Softmax Thresholding
Dec 12, 2025Abstract:The growing demand for long-context inference capabilities in Large Language Models (LLMs) has intensified the computational and memory bottlenecks inherent to the standard attention mechanism. To address this challenge, we introduce BLASST, a drop-in sparse attention method that dynamically prunes the attention matrix without any pre-computation or proxy scores. Our method uses a fixed threshold and existing information from online softmax to identify negligible attention scores, skipping softmax computation, Value block loading, and the subsequent matrix multiplication. This fits seamlessly into existing FlashAttention kernel designs with negligible latency overhead. The approach is applicable to both prefill and decode stages across all attention variants (MHA, GQA, MQA, and MLA), providing a unified solution for accelerating long-context inference. We develop an automated calibration procedure that reveals a simple inverse relationship between optimal threshold and context length, enabling robust deployment across diverse scenarios. Maintaining high accuracy, we demonstrate a 1.62x speedup for prefill at 74.7% sparsity and a 1.48x speedup for decode at 73.2% sparsity on modern GPUs. Furthermore, we explore sparsity-aware training as a natural extension, showing that models can be trained to be inherently more robust to sparse attention patterns, pushing the accuracy-sparsity frontier even further.
Give Me FP32 or Give Me Death? Challenges and Solutions for Reproducible Reasoning
Jun 11, 2025Abstract:Large Language Models (LLMs) are now integral across various domains and have demonstrated impressive performance. Progress, however, rests on the premise that benchmark scores are both accurate and reproducible. We demonstrate that the reproducibility of LLM performance is fragile: changing system configuration such as evaluation batch size, GPU count, and GPU version can introduce significant difference in the generated responses. This issue is especially pronounced in reasoning models, where minor rounding differences in early tokens can cascade into divergent chains of thought, ultimately affecting accuracy. For instance, under bfloat16 precision with greedy decoding, a reasoning model like DeepSeek-R1-Distill-Qwen-7B can exhibit up to 9% variation in accuracy and 9,000 tokens difference in response length due to differences in GPU count, type, and evaluation batch size. We trace the root cause of this variability to the non-associative nature of floating-point arithmetic under limited numerical precision. This work presents the first systematic investigation into how numerical precision affects reproducibility in LLM inference. Through carefully controlled experiments across various hardware, software, and precision settings, we quantify when and how model outputs diverge. Our analysis reveals that floating-point precision -- while critical for reproducibility -- is often neglected in evaluation practices. Inspired by this, we develop a lightweight inference pipeline, dubbed LayerCast, that stores weights in 16-bit precision but performs all computations in FP32, balancing memory efficiency with numerical stability. Code is available at https://github.com/nanomaoli/llm_reproducibility.
AutoL2S: Auto Long-Short Reasoning for Efficient Large Language Models
May 28, 2025Abstract:The reasoning-capable large language models (LLMs) demonstrate strong performance on complex reasoning tasks but often suffer from overthinking, generating unnecessarily long chain-of-thought (CoT) reasoning paths for easy reasoning questions, thereby increasing inference cost and latency. Recent approaches attempt to address this challenge by manually deciding when to apply long or short reasoning. However, they lack the flexibility to adapt CoT length dynamically based on question complexity. In this paper, we propose Auto Long-Short Reasoning (AutoL2S), a dynamic and model-agnostic framework that enables LLMs to dynamically compress their generated reasoning path based on the complexity of the reasoning question. AutoL2S enables a learned paradigm, in which LLMs themselves can decide when longer reasoning is necessary and when shorter reasoning suffices, by training on data annotated with our proposed method, which includes both long and short CoT paths and a special <EASY> token. We then use <EASY> token to indicate when the model can skip generating lengthy CoT reasoning. This proposed annotation strategy can enhance the LLMs' ability to generate shorter CoT reasoning paths with improved quality after training. Extensive evaluation results show that AutoL2S reduces the length of reasoning generation by up to 57% without compromising performance, demonstrating the effectiveness of AutoL2S for scalable and efficient LLM reasoning.
70% Size, 100% Accuracy: Lossless LLM Compression for Efficient GPU Inference via Dynamic-Length Float
Apr 15, 2025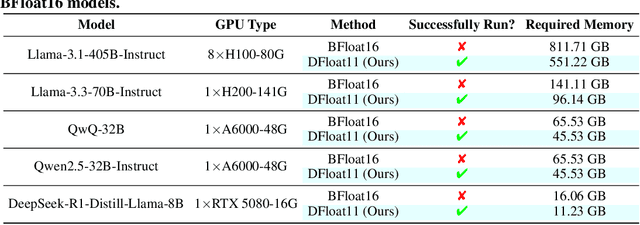
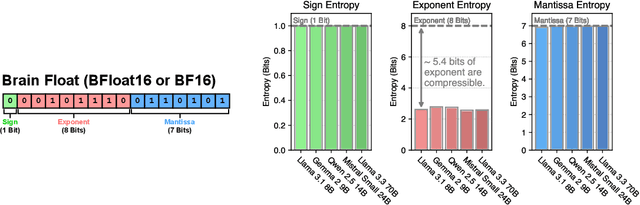
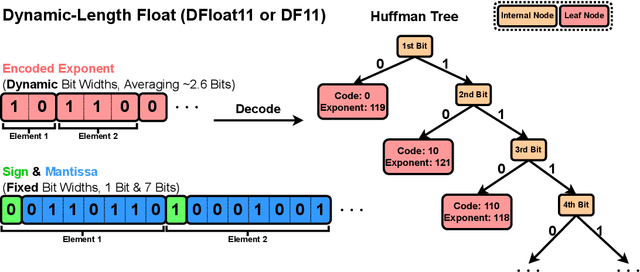
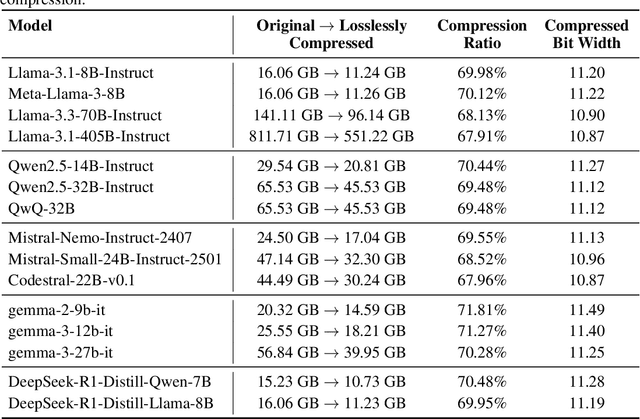
Abstract:Large Language Models (LLMs) have grown rapidly in size, creating significant challenges for efficient deployment on resource-constrained hardware. In this paper, we introduce Dynamic-Length Float (DFloat11), a lossless compression framework that reduces LLM size by 30% while preserving outputs that are bit-for-bit identical to the original model. DFloat11 is motivated by the low entropy in the BFloat16 weight representation of LLMs, which reveals significant inefficiency in existing storage format. By applying entropy coding, DFloat11 assigns dynamic-length encodings to weights based on frequency, achieving near information-optimal compression without any loss of precision. To facilitate efficient inference with dynamic-length encodings, we develop a custom GPU kernel for fast online decompression. Our design incorporates the following: (i) decomposition of memory-intensive lookup tables (LUTs) into compact LUTs that fit in GPU SRAM, (ii) a two-phase kernel for coordinating thread read/write positions using lightweight auxiliary variables, and (iii) transformer-block-level decompression to minimize latency. Experiments on recent models, including Llama-3.1, Qwen-2.5, and Gemma-3, validates our hypothesis that DFloat11 achieves around 30% model size reduction while preserving bit-for-bit exact outputs. Compared to a potential alternative of offloading parts of an uncompressed model to the CPU to meet memory constraints, DFloat11 achieves 1.9-38.8x higher throughput in token generation. With a fixed GPU memory budget, DFloat11 enables 5.3-13.17x longer context lengths than uncompressed models. Notably, our method enables lossless inference of Llama-3.1-405B, an 810GB model, on a single node equipped with 8x80GB GPUs. Our code and models are available at https://github.com/LeanModels/DFloat11.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge