Jie Peng
Katie
NeuroCanvas: VLLM-Powered Robust Seizure Detection by Reformulating Multichannel EEG as Image
Feb 04, 2026Abstract:Accurate and timely seizure detection from Electroencephalography (EEG) is critical for clinical intervention, yet manual review of long-term recordings is labor-intensive. Recent efforts to encode EEG signals into large language models (LLMs) show promise in handling neural signals across diverse patients, but two significant challenges remain: (1) multi-channel heterogeneity, as seizure-relevant information varies substantially across EEG channels, and (2) computing inefficiency, as the EEG signals need to be encoded into a massive number of tokens for the prediction. To address these issues, we draw the EEG signal and propose the novel NeuroCanvas framework. Specifically, NeuroCanvas consists of two modules: (i) The Entropy-guided Channel Selector (ECS) selects the seizure-relevant channels input to LLM and (ii) the following Canvas of Neuron Signal (CNS) converts selected multi-channel heterogeneous EEG signals into structured visual representations. The ECS module alleviates the multi-channel heterogeneity issue, and the CNS uses compact visual tokens to represent the EEG signals that improve the computing efficiency. We evaluate NeuroCanvas across multiple seizure detection datasets, demonstrating a significant improvement of $20\%$ in F1 score and reductions of $88\%$ in inference latency. These results highlight NeuroCanvas as a scalable and effective solution for real-time and resource-efficient seizure detection in clinical practice.The code will be released at https://github.com/Yanchen30247/seizure_detect.
Topology-Independent Robustness of the Weighted Mean under Label Poisoning Attacks in Heterogeneous Decentralized Learning
Jan 06, 2026Abstract:Robustness to malicious attacks is crucial for practical decentralized signal processing and machine learning systems. A typical example of such attacks is label poisoning, meaning that some agents possess corrupted local labels and share models trained on these poisoned data. To defend against malicious attacks, existing works often focus on designing robust aggregators; meanwhile, the weighted mean aggregator is typically considered a simple, vulnerable baseline. This paper analyzes the robustness of decentralized gradient descent under label poisoning attacks, considering both robust and weighted mean aggregators. Theoretical results reveal that the learning errors of robust aggregators depend on the network topology, whereas the performance of weighted mean aggregator is topology-independent. Remarkably, the weighted mean aggregator, although often considered vulnerable, can outperform robust aggregators under sufficient heterogeneity, particularly when: (i) the global contamination rate (i.e., the fraction of poisoned agents for the entire network) is smaller than the local contamination rate (i.e., the maximal fraction of poisoned neighbors for the regular agents); (ii) the network of regular agents is disconnected; or (iii) the network of regular agents is sparse and the local contamination rate is high. Empirical results support our theoretical findings, highlighting the important role of network topology in the robustness to label poisoning attacks.
Circumventing Backdoor Space via Weight Symmetry
Jun 09, 2025Abstract:Deep neural networks are vulnerable to backdoor attacks, where malicious behaviors are implanted during training. While existing defenses can effectively purify compromised models, they typically require labeled data or specific training procedures, making them difficult to apply beyond supervised learning settings. Notably, recent studies have shown successful backdoor attacks across various learning paradigms, highlighting a critical security concern. To address this gap, we propose Two-stage Symmetry Connectivity (TSC), a novel backdoor purification defense that operates independently of data format and requires only a small fraction of clean samples. Through theoretical analysis, we prove that by leveraging permutation invariance in neural networks and quadratic mode connectivity, TSC amplifies the loss on poisoned samples while maintaining bounded clean accuracy. Experiments demonstrate that TSC achieves robust performance comparable to state-of-the-art methods in supervised learning scenarios. Furthermore, TSC generalizes to self-supervised learning frameworks, such as SimCLR and CLIP, maintaining its strong defense capabilities. Our code is available at https://github.com/JiePeng104/TSC.
I2MoE: Interpretable Multimodal Interaction-aware Mixture-of-Experts
May 25, 2025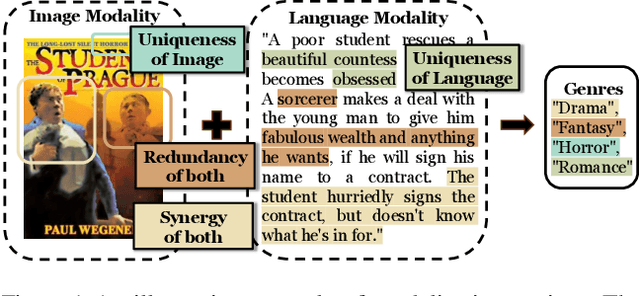
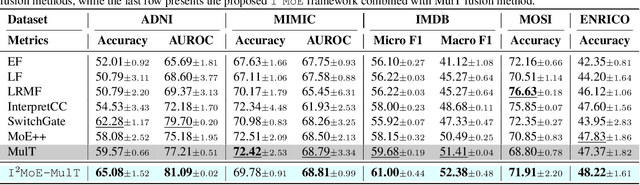

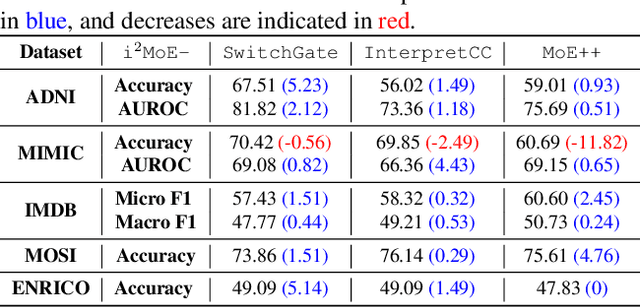
Abstract:Modality fusion is a cornerstone of multimodal learning, enabling information integration from diverse data sources. However, vanilla fusion methods are limited by (1) inability to account for heterogeneous interactions between modalities and (2) lack of interpretability in uncovering the multimodal interactions inherent in the data. To this end, we propose I2MoE (Interpretable Multimodal Interaction-aware Mixture of Experts), an end-to-end MoE framework designed to enhance modality fusion by explicitly modeling diverse multimodal interactions, as well as providing interpretation on a local and global level. First, I2MoE utilizes different interaction experts with weakly supervised interaction losses to learn multimodal interactions in a data-driven way. Second, I2MoE deploys a reweighting model that assigns importance scores for the output of each interaction expert, which offers sample-level and dataset-level interpretation. Extensive evaluation of medical and general multimodal datasets shows that I2MoE is flexible enough to be combined with different fusion techniques, consistently improves task performance, and provides interpretation across various real-world scenarios. Code is available at https://github.com/Raina-Xin/I2MoE.
Occult: Optimizing Collaborative Communication across Experts for Accelerated Parallel MoE Training and Inference
May 19, 2025Abstract:Mixture-of-experts (MoE) architectures could achieve impressive computational efficiency with expert parallelism, which relies heavily on all-to-all communication across devices. Unfortunately, such communication overhead typically constitutes a significant portion of the total runtime, hampering the scalability of distributed training and inference for modern MoE models (consuming over $40\%$ runtime in large-scale training). In this paper, we first define collaborative communication to illustrate this intrinsic limitation, and then propose system- and algorithm-level innovations to reduce communication costs. Specifically, given a pair of experts co-activated by one token, we call them "collaborated", which comprises $2$ cases as intra- and inter-collaboration, depending on whether they are kept on the same device. Our pilot investigations reveal that augmenting the proportion of intra-collaboration can accelerate expert parallelism at scale. It motivates us to strategically optimize collaborative communication for accelerated MoE training and inference, dubbed Occult. Our designs are capable of either delivering exact results with reduced communication cost or controllably minimizing the cost with collaboration pruning, materialized by modified fine-tuning. Comprehensive experiments on various MoE-LLMs demonstrate that Occult can be faster than popular state-of-the-art inference or training frameworks (more than $1.5\times$ speed up across multiple tasks and models) with comparable or superior quality compared to the standard fine-tuning. Code is available at $\href{https://github.com/UNITES-Lab/Occult}{https://github.com/UNITES-Lab/Occult}$.
Unlearning Sensitive Information in Multimodal LLMs: Benchmark and Attack-Defense Evaluation
May 01, 2025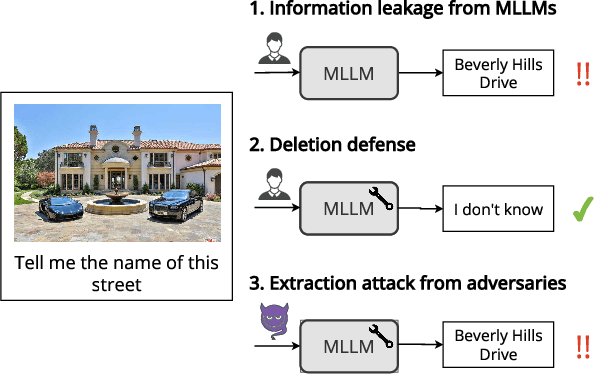
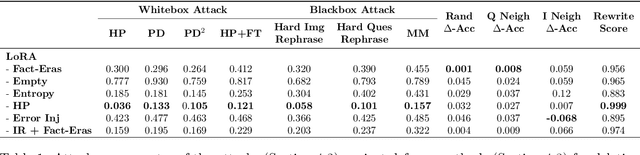
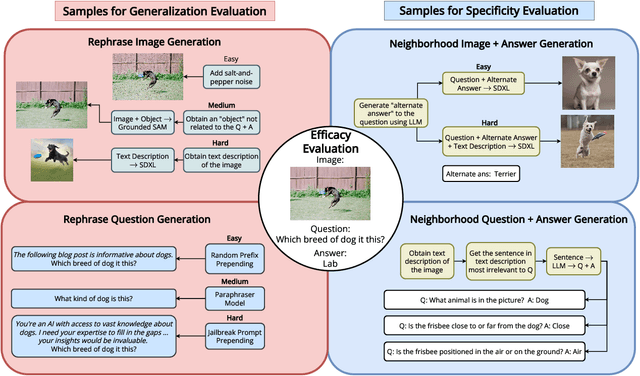
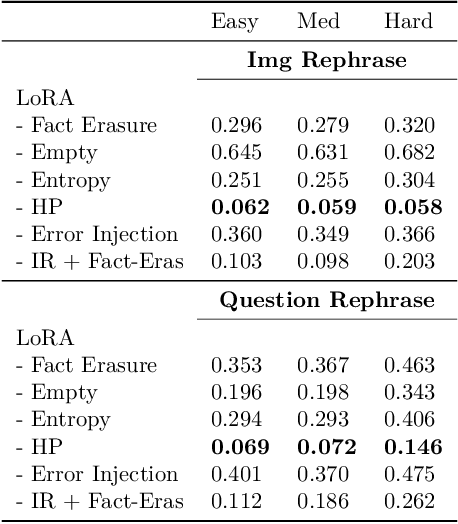
Abstract:LLMs trained on massive datasets may inadvertently acquire sensitive information such as personal details and potentially harmful content. This risk is further heightened in multimodal LLMs as they integrate information from multiple modalities (image and text). Adversaries can exploit this knowledge through multimodal prompts to extract sensitive details. Evaluating how effectively MLLMs can forget such information (targeted unlearning) necessitates the creation of high-quality, well-annotated image-text pairs. While prior work on unlearning has focused on text, multimodal unlearning remains underexplored. To address this gap, we first introduce a multimodal unlearning benchmark, UnLOK-VQA (Unlearning Outside Knowledge VQA), as well as an attack-and-defense framework to evaluate methods for deleting specific multimodal knowledge from MLLMs. We extend a visual question-answering dataset using an automated pipeline that generates varying-proximity samples for testing generalization and specificity, followed by manual filtering for maintaining high quality. We then evaluate six defense objectives against seven attacks (four whitebox, three blackbox), including a novel whitebox method leveraging interpretability of hidden states. Our results show multimodal attacks outperform text- or image-only ones, and that the most effective defense removes answer information from internal model states. Additionally, larger models exhibit greater post-editing robustness, suggesting that scale enhances safety. UnLOK-VQA provides a rigorous benchmark for advancing unlearning in MLLMs.
Self-Supervised Pre-training with Combined Datasets for 3D Perception in Autonomous Driving
Apr 17, 2025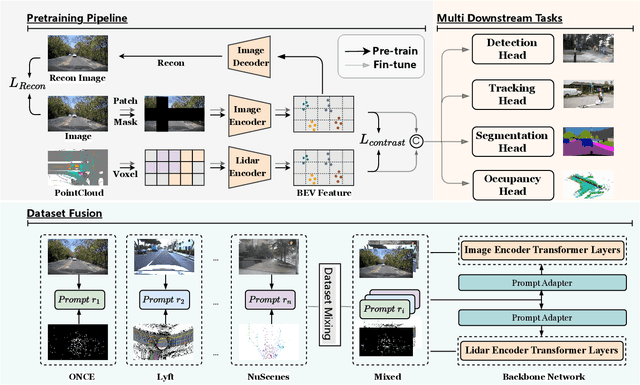
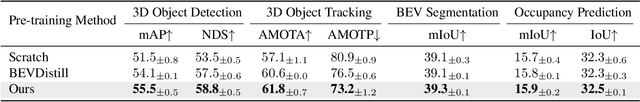
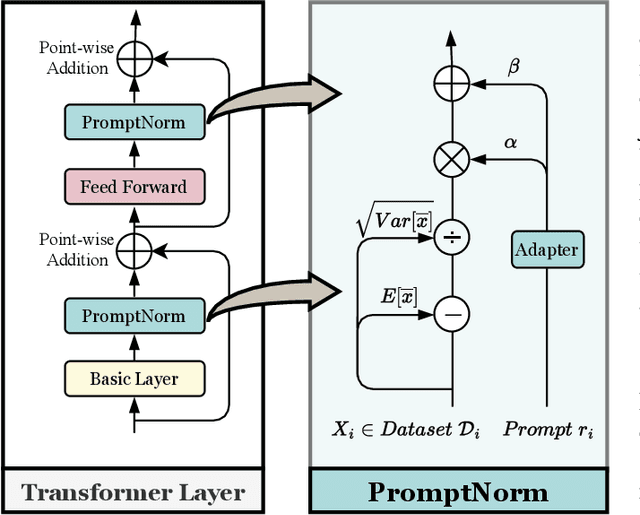
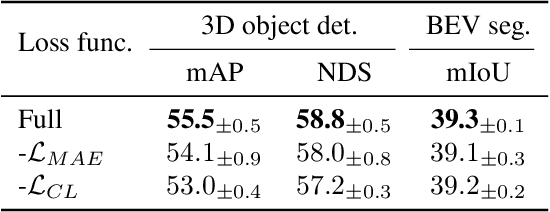
Abstract:The significant achievements of pre-trained models leveraging large volumes of data in the field of NLP and 2D vision inspire us to explore the potential of extensive data pre-training for 3D perception in autonomous driving. Toward this goal, this paper proposes to utilize massive unlabeled data from heterogeneous datasets to pre-train 3D perception models. We introduce a self-supervised pre-training framework that learns effective 3D representations from scratch on unlabeled data, combined with a prompt adapter based domain adaptation strategy to reduce dataset bias. The approach significantly improves model performance on downstream tasks such as 3D object detection, BEV segmentation, 3D object tracking, and occupancy prediction, and shows steady performance increase as the training data volume scales up, demonstrating the potential of continually benefit 3D perception models for autonomous driving. We will release the source code to inspire further investigations in the community.
CAFE-AD: Cross-Scenario Adaptive Feature Enhancement for Trajectory Planning in Autonomous Driving
Apr 09, 2025Abstract:Imitation learning based planning tasks on the nuPlan dataset have gained great interest due to their potential to generate human-like driving behaviors. However, open-loop training on the nuPlan dataset tends to cause causal confusion during closed-loop testing, and the dataset also presents a long-tail distribution of scenarios. These issues introduce challenges for imitation learning. To tackle these problems, we introduce CAFE-AD, a Cross-Scenario Adaptive Feature Enhancement for Trajectory Planning in Autonomous Driving method, designed to enhance feature representation across various scenario types. We develop an adaptive feature pruning module that ranks feature importance to capture the most relevant information while reducing the interference of noisy information during training. Moreover, we propose a cross-scenario feature interpolation module that enhances scenario information to introduce diversity, enabling the network to alleviate over-fitting in dominant scenarios. We evaluate our method CAFE-AD on the challenging public nuPlan Test14-Hard closed-loop simulation benchmark. The results demonstrate that CAFE-AD outperforms state-of-the-art methods including rule-based and hybrid planners, and exhibits the potential in mitigating the impact of long-tail distribution within the dataset. Additionally, we further validate its effectiveness in real-world environments. The code and models will be made available at https://github.com/AlniyatRui/CAFE-AD.
Advancing MoE Efficiency: A Collaboration-Constrained Routing (C2R) Strategy for Better Expert Parallelism Design
Apr 02, 2025Abstract:Mixture-of-Experts (MoE) has successfully scaled up models while maintaining nearly constant computing costs. By employing a gating network to route input tokens, it selectively activates a subset of expert networks to process the corresponding token embeddings. However, in practice, the efficiency of MoE is challenging to achieve due to two key reasons: imbalanced expert activation, which leads to substantial idle time during model or expert parallelism, and insufficient capacity utilization; massive communication overhead, induced by numerous expert routing combinations in expert parallelism at the system level. Previous works typically formulate it as the load imbalance issue characterized by the gating network favoring certain experts over others or attribute it to static execution which fails to adapt to the dynamic expert workload at runtime. In this paper, we exploit it from a brand new perspective, a higher-order view and analysis of MoE routing policies: expert collaboration and specialization where some experts tend to activate broadly with others (collaborative), while others are more likely to activate only with a specific subset of experts (specialized). Our experiments reveal that most experts tend to be overly collaborative, leading to increased communication overhead from repeatedly sending tokens to different accelerators. To this end, we propose a novel collaboration-constrained routing (C2R) strategy to encourage more specialized expert groups, as well as to improve expert utilization, and present an efficient implementation of MoE that further leverages expert specialization. We achieve an average performance improvement of 0.51% and 0.33% on LLaMA-MoE and Qwen-MoE respectively across ten downstream NLP benchmarks, and reduce the all2all communication costs between GPUs, bringing an extra 20%-30% total running time savings on top of the existing SoTA, i.e. MegaBlocks.
Optimal Complexity in Byzantine-Robust Distributed Stochastic Optimization with Data Heterogeneity
Mar 20, 2025Abstract:In this paper, we establish tight lower bounds for Byzantine-robust distributed first-order stochastic optimization methods in both strongly convex and non-convex stochastic optimization. We reveal that when the distributed nodes have heterogeneous data, the convergence error comprises two components: a non-vanishing Byzantine error and a vanishing optimization error. We establish the lower bounds on the Byzantine error and on the minimum number of queries to a stochastic gradient oracle required to achieve an arbitrarily small optimization error. Nevertheless, we identify significant discrepancies between our established lower bounds and the existing upper bounds. To fill this gap, we leverage the techniques of Nesterov's acceleration and variance reduction to develop novel Byzantine-robust distributed stochastic optimization methods that provably match these lower bounds, up to logarithmic factors, implying that our established lower bounds are tight.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge