Laila Rasmy
Advancing Pancreatic Cancer Prediction with a Next Visit Token Prediction Head on top of Med-BERT
Jan 03, 2025



Abstract:Background: Recently, numerous foundation models pretrained on extensive data have demonstrated efficacy in disease prediction using Electronic Health Records (EHRs). However, there remains some unanswered questions on how to best utilize such models especially with very small fine-tuning cohorts. Methods: We utilized Med-BERT, an EHR-specific foundation model, and reformulated the disease binary prediction task into a token prediction task and a next visit mask token prediction task to align with Med-BERT's pretraining task format in order to improve the accuracy of pancreatic cancer (PaCa) prediction in both few-shot and fully supervised settings. Results: The reformulation of the task into a token prediction task, referred to as Med-BERT-Sum, demonstrates slightly superior performance in both few-shot scenarios and larger data samples. Furthermore, reformulating the prediction task as a Next Visit Mask Token Prediction task (Med-BERT-Mask) significantly outperforms the conventional Binary Classification (BC) prediction task (Med-BERT-BC) by 3% to 7% in few-shot scenarios with data sizes ranging from 10 to 500 samples. These findings highlight that aligning the downstream task with Med-BERT's pretraining objectives substantially enhances the model's predictive capabilities, thereby improving its effectiveness in predicting both rare and common diseases. Conclusion: Reformatting disease prediction tasks to align with the pretraining of foundation models enhances prediction accuracy, leading to earlier detection and timely intervention. This approach improves treatment effectiveness, survival rates, and overall patient outcomes for PaCa and potentially other cancers.
Prompting Large Language Models for Clinical Temporal Relation Extraction
Dec 04, 2024Abstract:Objective: This paper aims to prompt large language models (LLMs) for clinical temporal relation extraction (CTRE) in both few-shot and fully supervised settings. Materials and Methods: This study utilizes four LLMs: Encoder-based GatorTron-Base (345M)/Large (8.9B); Decoder-based LLaMA3-8B/MeLLaMA-13B. We developed full (FFT) and parameter-efficient (PEFT) fine-tuning strategies and evaluated these strategies on the 2012 i2b2 CTRE task. We explored four fine-tuning strategies for GatorTron-Base: (1) Standard Fine-Tuning, (2) Hard-Prompting with Unfrozen LLMs, (3) Soft-Prompting with Frozen LLMs, and (4) Low-Rank Adaptation (LoRA) with Frozen LLMs. For GatorTron-Large, we assessed two PEFT strategies-Soft-Prompting and LoRA with Frozen LLMs-leveraging Quantization techniques. Additionally, LLaMA3-8B and MeLLaMA-13B employed two PEFT strategies: LoRA strategy with Quantization (QLoRA) applied to Frozen LLMs using instruction tuning and standard fine-tuning. Results: Under fully supervised settings, Hard-Prompting with Unfrozen GatorTron-Base achieved the highest F1 score (89.54%), surpassing the SOTA model (85.70%) by 3.74%. Additionally, two variants of QLoRA adapted to GatorTron-Large and Standard Fine-Tuning of GatorTron-Base exceeded the SOTA model by 2.36%, 1.88%, and 0.25%, respectively. Decoder-based models with frozen parameters outperformed their Encoder-based counterparts in this setting; however, the trend reversed in few-shot scenarios. Discussions and Conclusions: This study presented new methods that significantly improved CTRE performance, benefiting downstream tasks reliant on CTRE systems. The findings underscore the importance of selecting appropriate models and fine-tuning strategies based on task requirements and data availability. Future work will explore larger models and broader CTRE applications.
PheME: A deep ensemble framework for improving phenotype prediction from multi-modal data
Mar 19, 2023Abstract:Detailed phenotype information is fundamental to accurate diagnosis and risk estimation of diseases. As a rich source of phenotype information, electronic health records (EHRs) promise to empower diagnostic variant interpretation. However, how to accurately and efficiently extract phenotypes from the heterogeneous EHR data remains a challenge. In this work, we present PheME, an Ensemble framework using Multi-modality data of structured EHRs and unstructured clinical notes for accurate Phenotype prediction. Firstly, we employ multiple deep neural networks to learn reliable representations from the sparse structured EHR data and redundant clinical notes. A multi-modal model then aligns multi-modal features onto the same latent space to predict phenotypes. Secondly, we leverage ensemble learning to combine outputs from single-modal models and multi-modal models to improve phenotype predictions. We choose seven diseases to evaluate the phenotyping performance of the proposed framework. Experimental results show that using multi-modal data significantly improves phenotype prediction in all diseases, the proposed ensemble learning framework can further boost the performance.
Simple Recurrent Neural Networks is all we need for clinical events predictions using EHR data
Oct 03, 2021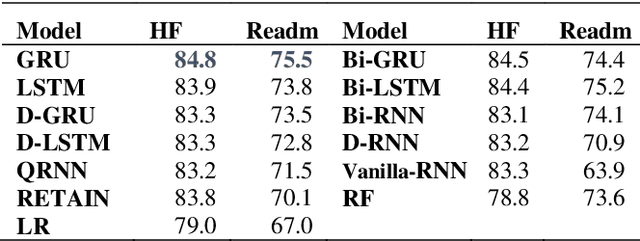
Abstract:Recently, there is great interest to investigate the application of deep learning models for the prediction of clinical events using electronic health records (EHR) data. In EHR data, a patient's history is often represented as a sequence of visits, and each visit contains multiple events. As a result, deep learning models developed for sequence modeling, like recurrent neural networks (RNNs) are common architecture for EHR-based clinical events predictive models. While a large variety of RNN models were proposed in the literature, it is unclear if complex architecture innovations will offer superior predictive performance. In order to move this field forward, a rigorous evaluation of various methods is needed. In this study, we conducted a thorough benchmark of RNN architectures in modeling EHR data. We used two prediction tasks: the risk for developing heart failure and the risk of early readmission for inpatient hospitalization. We found that simple gated RNN models, including GRUs and LSTMs, often offer competitive results when properly tuned with Bayesian Optimization, which is in line with similar to findings in the natural language processing (NLP) domain. For reproducibility, Our codebase is shared at https://github.com/ZhiGroup/pytorch_ehr.
Med-BERT: pre-trained contextualized embeddings on large-scale structured electronic health records for disease prediction
May 22, 2020



Abstract:Deep learning (DL) based predictive models from electronic health records (EHR) deliver impressive performance in many clinical tasks. Large training cohorts, however, are often required to achieve high accuracy, hindering the adoption of DL-based models in scenarios with limited training data size. Recently, bidirectional encoder representations from transformers (BERT) and related models have achieved tremendous successes in the natural language processing domain. The pre-training of BERT on a very large training corpus generates contextualized embeddings that can boost the performance of models trained on smaller datasets. We propose Med-BERT, which adapts the BERT framework for pre-training contextualized embedding models on structured diagnosis data from 28,490,650 patients EHR dataset. Fine-tuning experiments are conducted on two disease-prediction tasks: (1) prediction of heart failure in patients with diabetes and (2) prediction of pancreatic cancer from two clinical databases. Med-BERT substantially improves prediction accuracy, boosting the area under receiver operating characteristics curve (AUC) by 2.02-7.12%. In particular, pre-trained Med-BERT substantially improves the performance of tasks with very small fine-tuning training sets (300-500 samples) boosting the AUC by more than 20% or equivalent to the AUC of 10 times larger training set. We believe that Med-BERT will benefit disease-prediction studies with small local training datasets, reduce data collection expenses, and accelerate the pace of artificial intelligence aided healthcare.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge