Jianfu Li
Prompting Large Language Models for Clinical Temporal Relation Extraction
Dec 04, 2024Abstract:Objective: This paper aims to prompt large language models (LLMs) for clinical temporal relation extraction (CTRE) in both few-shot and fully supervised settings. Materials and Methods: This study utilizes four LLMs: Encoder-based GatorTron-Base (345M)/Large (8.9B); Decoder-based LLaMA3-8B/MeLLaMA-13B. We developed full (FFT) and parameter-efficient (PEFT) fine-tuning strategies and evaluated these strategies on the 2012 i2b2 CTRE task. We explored four fine-tuning strategies for GatorTron-Base: (1) Standard Fine-Tuning, (2) Hard-Prompting with Unfrozen LLMs, (3) Soft-Prompting with Frozen LLMs, and (4) Low-Rank Adaptation (LoRA) with Frozen LLMs. For GatorTron-Large, we assessed two PEFT strategies-Soft-Prompting and LoRA with Frozen LLMs-leveraging Quantization techniques. Additionally, LLaMA3-8B and MeLLaMA-13B employed two PEFT strategies: LoRA strategy with Quantization (QLoRA) applied to Frozen LLMs using instruction tuning and standard fine-tuning. Results: Under fully supervised settings, Hard-Prompting with Unfrozen GatorTron-Base achieved the highest F1 score (89.54%), surpassing the SOTA model (85.70%) by 3.74%. Additionally, two variants of QLoRA adapted to GatorTron-Large and Standard Fine-Tuning of GatorTron-Base exceeded the SOTA model by 2.36%, 1.88%, and 0.25%, respectively. Decoder-based models with frozen parameters outperformed their Encoder-based counterparts in this setting; however, the trend reversed in few-shot scenarios. Discussions and Conclusions: This study presented new methods that significantly improved CTRE performance, benefiting downstream tasks reliant on CTRE systems. The findings underscore the importance of selecting appropriate models and fine-tuning strategies based on task requirements and data availability. Future work will explore larger models and broader CTRE applications.
Improving Entity Recognition Using Ensembles of Deep Learning and Fine-tuned Large Language Models: A Case Study on Adverse Event Extraction from Multiple Sources
Jun 26, 2024Abstract:Adverse event (AE) extraction following COVID-19 vaccines from text data is crucial for monitoring and analyzing the safety profiles of immunizations. Traditional deep learning models are adept at learning intricate feature representations and dependencies in sequential data, but often require extensive labeled data. In contrast, large language models (LLMs) excel in understanding contextual information, but exhibit unstable performance on named entity recognition tasks, possibly due to their broad but unspecific training. This study aims to evaluate the effectiveness of LLMs and traditional deep learning models in AE extraction, and to assess the impact of ensembling these models on performance. In this study, we utilized reports and posts from the VAERS (n=621), Twitter (n=9,133), and Reddit (n=131) as our corpora. Our goal was to extract three types of entities: "vaccine", "shot", and "ae". We explored and fine-tuned (except GPT-4) multiple LLMs, including GPT-2, GPT-3.5, GPT-4, and Llama-2, as well as traditional deep learning models like RNN and BioBERT. To enhance performance, we created ensembles of the three models with the best performance. For evaluation, we used strict and relaxed F1 scores to evaluate the performance for each entity type, and micro-average F1 was used to assess the overall performance. The ensemble model achieved the highest performance in "vaccine", "shot", and "ae" with strict F1-scores of 0.878, 0.930, and 0.925, respectively, along with a micro-average score of 0.903. In conclusion, this study demonstrates the effectiveness and robustness of ensembling fine-tuned traditional deep learning models and LLMs, for extracting AE-related information. This study contributes to the advancement of biomedical natural language processing, providing valuable insights into improving AE extraction from text data for pharmacovigilance and public health surveillance.
Relation Extraction Using Large Language Models: A Case Study on Acupuncture Point Locations
Apr 15, 2024Abstract:In acupuncture therapy, the accurate location of acupoints is essential for its effectiveness. The advanced language understanding capabilities of large language models (LLMs) like Generative Pre-trained Transformers (GPT) present a significant opportunity for extracting relations related to acupoint locations from textual knowledge sources. This study aims to compare the performance of GPT with traditional deep learning models (Long Short-Term Memory (LSTM) and Bidirectional Encoder Representations from Transformers for Biomedical Text Mining (BioBERT)) in extracting acupoint-related location relations and assess the impact of pretraining and fine-tuning on GPT's performance. We utilized the World Health Organization Standard Acupuncture Point Locations in the Western Pacific Region (WHO Standard) as our corpus, which consists of descriptions of 361 acupoints. Five types of relations ('direction_of,' 'distance_of,' 'part_of,' 'near_acupoint,' and 'located_near') (n= 3,174) between acupoints were annotated. Five models were compared: BioBERT, LSTM, pre-trained GPT-3.5, fine-tuned GPT-3.5, as well as pre-trained GPT-4. Performance metrics included micro-average exact match precision, recall, and F1 scores. Our results demonstrate that fine-tuned GPT-3.5 consistently outperformed other models in F1 scores across all relation types. Overall, it achieved the highest micro-average F1 score of 0.92. This study underscores the effectiveness of LLMs like GPT in extracting relations related to acupoint locations, with implications for accurately modeling acupuncture knowledge and promoting standard implementation in acupuncture training and practice. The findings also contribute to advancing informatics applications in traditional and complementary medicine, showcasing the potential of LLMs in natural language processing.
AE-GPT: Using Large Language Models to Extract Adverse Events from Surveillance Reports-A Use Case with Influenza Vaccine Adverse Events
Sep 28, 2023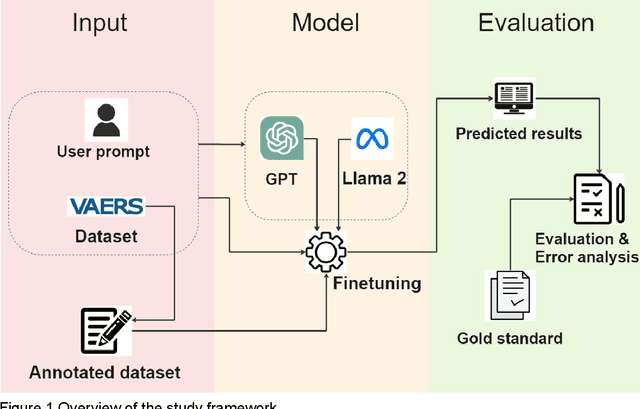
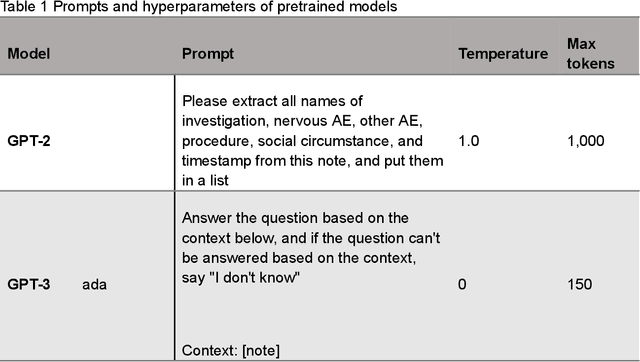


Abstract:Though Vaccines are instrumental in global health, mitigating infectious diseases and pandemic outbreaks, they can occasionally lead to adverse events (AEs). Recently, Large Language Models (LLMs) have shown promise in effectively identifying and cataloging AEs within clinical reports. Utilizing data from the Vaccine Adverse Event Reporting System (VAERS) from 1990 to 2016, this study particularly focuses on AEs to evaluate LLMs' capability for AE extraction. A variety of prevalent LLMs, including GPT-2, GPT-3 variants, GPT-4, and Llama 2, were evaluated using Influenza vaccine as a use case. The fine-tuned GPT 3.5 model (AE-GPT) stood out with a 0.704 averaged micro F1 score for strict match and 0.816 for relaxed match. The encouraging performance of the AE-GPT underscores LLMs' potential in processing medical data, indicating a significant stride towards advanced AE detection, thus presumably generalizable to other AE extraction tasks.
Zero-shot Clinical Entity Recognition using ChatGPT
Mar 29, 2023



Abstract:In this study, we investigated the potential of ChatGPT, a large language model developed by OpenAI, for the clinical named entity recognition task defined in the 2010 i2b2 challenge, in a zero-shot setting with two different prompt strategies. We compared its performance with GPT-3 in a similar zero-shot setting, as well as a fine-tuned BioClinicalBERT model using a set of synthetic clinical notes from MTSamples. Our findings revealed that ChatGPT outperformed GPT-3 in the zero-shot setting, with F1 scores of 0.418 (vs.0.250) and 0.620 (vs. 0.480) for exact- and relaxed-matching, respectively. Moreover, prompts affected ChatGPT's performance greatly, with relaxed-matching F1 scores of 0.628 vs.0.541 for two different prompt strategies. Although ChatGPT's performance was still lower than that of the supervised BioClinicalBERT model (i.e., relaxed-matching F1 scores of 0.628 vs. 0.870), our study demonstrates the great potential of ChatGPT for clinical NER tasks in a zero-shot setting, which is much more appealing as it does not require any annotation.
Robustly Pre-trained Neural Model for Direct Temporal Relation Extraction
Apr 13, 2020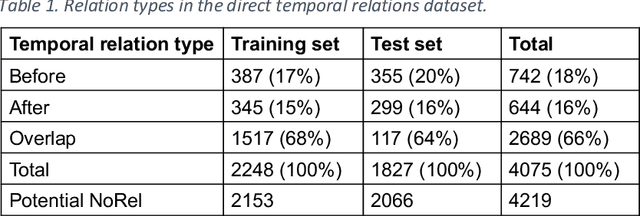
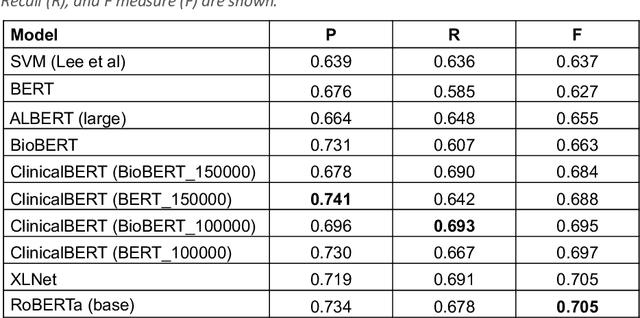
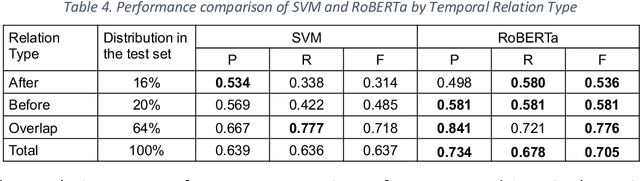
Abstract:Background: Identifying relationships between clinical events and temporal expressions is a key challenge in meaningfully analyzing clinical text for use in advanced AI applications. While previous studies exist, the state-of-the-art performance has significant room for improvement. Methods: We studied several variants of BERT (Bidirectional Encoder Representations using Transformers) some involving clinical domain customization and the others involving improved architecture and/or training strategies. We evaluated these methods using a direct temporal relations dataset which is a semantically focused subset of the 2012 i2b2 temporal relations challenge dataset. Results: Our results show that RoBERTa, which employs better pre-training strategies including using 10x larger corpus, has improved overall F measure by 0.0864 absolute score (on the 1.00 scale) and thus reducing the error rate by 24% relative to the previous state-of-the-art performance achieved with an SVM (support vector machine) model. Conclusion: Modern contextual language modeling neural networks, pre-trained on a large corpus, achieve impressive performance even on highly-nuanced clinical temporal relation tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge