Cui Tao
Early GVHD Prediction in Liver Transplantation via Multi-Modal Deep Learning on Imbalanced EHR Data
Nov 06, 2025Abstract:Graft-versus-host disease (GVHD) is a rare but often fatal complication in liver transplantation, with a very high mortality rate. By harnessing multi-modal deep learning methods to integrate heterogeneous and imbalanced electronic health records (EHR), we aim to advance early prediction of GVHD, paving the way for timely intervention and improved patient outcomes. In this study, we analyzed pre-transplant electronic health records (EHR) spanning the period before surgery for 2,100 liver transplantation patients, including 42 cases of graft-versus-host disease (GVHD), from a cohort treated at Mayo Clinic between 1992 and 2025. The dataset comprised four major modalities: patient demographics, laboratory tests, diagnoses, and medications. We developed a multi-modal deep learning framework that dynamically fuses these modalities, handles irregular records with missing values, and addresses extreme class imbalance through AUC-based optimization. The developed framework outperforms all single-modal and multi-modal machine learning baselines, achieving an AUC of 0.836, an AUPRC of 0.157, a recall of 0.768, and a specificity of 0.803. It also demonstrates the effectiveness of our approach in capturing complementary information from different modalities, leading to improved performance. Our multi-modal deep learning framework substantially improves existing approaches for early GVHD prediction. By effectively addressing the challenges of heterogeneity and extreme class imbalance in real-world EHR, it achieves accurate early prediction. Our proposed multi-modal deep learning method demonstrates promising results for early prediction of a GVHD in liver transplantation, despite the challenge of extremely imbalanced EHR data.
ETT-CKGE: Efficient Task-driven Tokens for Continual Knowledge Graph Embedding
Jun 09, 2025Abstract:Continual Knowledge Graph Embedding (CKGE) seeks to integrate new knowledge while preserving past information. However, existing methods struggle with efficiency and scalability due to two key limitations: (1) suboptimal knowledge preservation between snapshots caused by manually designed node/relation importance scores that ignore graph dependencies relevant to the downstream task, and (2) computationally expensive graph traversal for node/relation importance calculation, leading to slow training and high memory overhead. To address these limitations, we introduce ETT-CKGE (Efficient, Task-driven, Tokens for Continual Knowledge Graph Embedding), a novel task-guided CKGE method that leverages efficient task-driven tokens for efficient and effective knowledge transfer between snapshots. Our method introduces a set of learnable tokens that directly capture task-relevant signals, eliminating the need for explicit node scoring or traversal. These tokens serve as consistent and reusable guidance across snapshots, enabling efficient token-masked embedding alignment between snapshots. Importantly, knowledge transfer is achieved through simple matrix operations, significantly reducing training time and memory usage. Extensive experiments across six benchmark datasets demonstrate that ETT-CKGE consistently achieves superior or competitive predictive performance, while substantially improving training efficiency and scalability compared to state-of-the-art CKGE methods. The code is available at: https://github.com/lijingzhu1/ETT-CKGE/tree/main
LLM-Match: An Open-Sourced Patient Matching Model Based on Large Language Models and Retrieval-Augmented Generation
Mar 17, 2025Abstract:Patient matching is the process of linking patients to appropriate clinical trials by accurately identifying and matching their medical records with trial eligibility criteria. We propose LLM-Match, a novel framework for patient matching leveraging fine-tuned open-source large language models. Our approach consists of four key components. First, a retrieval-augmented generation (RAG) module extracts relevant patient context from a vast pool of electronic health records (EHRs). Second, a prompt generation module constructs input prompts by integrating trial eligibility criteria (both inclusion and exclusion criteria), patient context, and system instructions. Third, a fine-tuning module with a classification head optimizes the model parameters using structured prompts and ground-truth labels. Fourth, an evaluation module assesses the fine-tuned model's performance on the testing datasets. We evaluated LLM-Match on four open datasets, n2c2, SIGIR, TREC 2021, and TREC 2022, using open-source models, comparing it against TrialGPT, Zero-Shot, and GPT-4-based closed models. LLM-Match outperformed all baselines.
Advancing Pancreatic Cancer Prediction with a Next Visit Token Prediction Head on top of Med-BERT
Jan 03, 2025



Abstract:Background: Recently, numerous foundation models pretrained on extensive data have demonstrated efficacy in disease prediction using Electronic Health Records (EHRs). However, there remains some unanswered questions on how to best utilize such models especially with very small fine-tuning cohorts. Methods: We utilized Med-BERT, an EHR-specific foundation model, and reformulated the disease binary prediction task into a token prediction task and a next visit mask token prediction task to align with Med-BERT's pretraining task format in order to improve the accuracy of pancreatic cancer (PaCa) prediction in both few-shot and fully supervised settings. Results: The reformulation of the task into a token prediction task, referred to as Med-BERT-Sum, demonstrates slightly superior performance in both few-shot scenarios and larger data samples. Furthermore, reformulating the prediction task as a Next Visit Mask Token Prediction task (Med-BERT-Mask) significantly outperforms the conventional Binary Classification (BC) prediction task (Med-BERT-BC) by 3% to 7% in few-shot scenarios with data sizes ranging from 10 to 500 samples. These findings highlight that aligning the downstream task with Med-BERT's pretraining objectives substantially enhances the model's predictive capabilities, thereby improving its effectiveness in predicting both rare and common diseases. Conclusion: Reformatting disease prediction tasks to align with the pretraining of foundation models enhances prediction accuracy, leading to earlier detection and timely intervention. This approach improves treatment effectiveness, survival rates, and overall patient outcomes for PaCa and potentially other cancers.
Prompting Large Language Models for Clinical Temporal Relation Extraction
Dec 04, 2024Abstract:Objective: This paper aims to prompt large language models (LLMs) for clinical temporal relation extraction (CTRE) in both few-shot and fully supervised settings. Materials and Methods: This study utilizes four LLMs: Encoder-based GatorTron-Base (345M)/Large (8.9B); Decoder-based LLaMA3-8B/MeLLaMA-13B. We developed full (FFT) and parameter-efficient (PEFT) fine-tuning strategies and evaluated these strategies on the 2012 i2b2 CTRE task. We explored four fine-tuning strategies for GatorTron-Base: (1) Standard Fine-Tuning, (2) Hard-Prompting with Unfrozen LLMs, (3) Soft-Prompting with Frozen LLMs, and (4) Low-Rank Adaptation (LoRA) with Frozen LLMs. For GatorTron-Large, we assessed two PEFT strategies-Soft-Prompting and LoRA with Frozen LLMs-leveraging Quantization techniques. Additionally, LLaMA3-8B and MeLLaMA-13B employed two PEFT strategies: LoRA strategy with Quantization (QLoRA) applied to Frozen LLMs using instruction tuning and standard fine-tuning. Results: Under fully supervised settings, Hard-Prompting with Unfrozen GatorTron-Base achieved the highest F1 score (89.54%), surpassing the SOTA model (85.70%) by 3.74%. Additionally, two variants of QLoRA adapted to GatorTron-Large and Standard Fine-Tuning of GatorTron-Base exceeded the SOTA model by 2.36%, 1.88%, and 0.25%, respectively. Decoder-based models with frozen parameters outperformed their Encoder-based counterparts in this setting; however, the trend reversed in few-shot scenarios. Discussions and Conclusions: This study presented new methods that significantly improved CTRE performance, benefiting downstream tasks reliant on CTRE systems. The findings underscore the importance of selecting appropriate models and fine-tuning strategies based on task requirements and data availability. Future work will explore larger models and broader CTRE applications.
A Comparative Study of Recent Large Language Models on Generating Hospital Discharge Summaries for Lung Cancer Patients
Nov 06, 2024


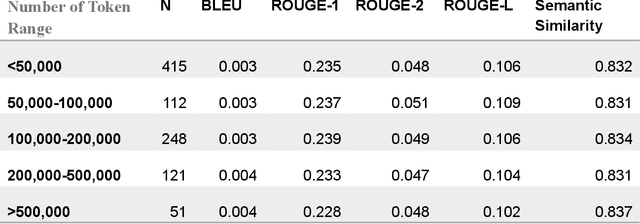
Abstract:Generating discharge summaries is a crucial yet time-consuming task in clinical practice, essential for conveying pertinent patient information and facilitating continuity of care. Recent advancements in large language models (LLMs) have significantly enhanced their capability in understanding and summarizing complex medical texts. This research aims to explore how LLMs can alleviate the burden of manual summarization, streamline workflow efficiencies, and support informed decision-making in healthcare settings. Clinical notes from a cohort of 1,099 lung cancer patients were utilized, with a subset of 50 patients for testing purposes, and 102 patients used for model fine-tuning. This study evaluates the performance of multiple LLMs, including GPT-3.5, GPT-4, GPT-4o, and LLaMA 3 8b, in generating discharge summaries. Evaluation metrics included token-level analysis (BLEU, ROUGE-1, ROUGE-2, ROUGE-L) and semantic similarity scores between model-generated summaries and physician-written gold standards. LLaMA 3 8b was further tested on clinical notes of varying lengths to examine the stability of its performance. The study found notable variations in summarization capabilities among LLMs. GPT-4o and fine-tuned LLaMA 3 demonstrated superior token-level evaluation metrics, while LLaMA 3 consistently produced concise summaries across different input lengths. Semantic similarity scores indicated GPT-4o and LLaMA 3 as leading models in capturing clinical relevance. This study contributes insights into the efficacy of LLMs for generating discharge summaries, highlighting LLaMA 3's robust performance in maintaining clarity and relevance across varying clinical contexts. These findings underscore the potential of automated summarization tools to enhance documentation precision and efficiency, ultimately improving patient care and operational capability in healthcare settings.
Improving Entity Recognition Using Ensembles of Deep Learning and Fine-tuned Large Language Models: A Case Study on Adverse Event Extraction from Multiple Sources
Jun 26, 2024Abstract:Adverse event (AE) extraction following COVID-19 vaccines from text data is crucial for monitoring and analyzing the safety profiles of immunizations. Traditional deep learning models are adept at learning intricate feature representations and dependencies in sequential data, but often require extensive labeled data. In contrast, large language models (LLMs) excel in understanding contextual information, but exhibit unstable performance on named entity recognition tasks, possibly due to their broad but unspecific training. This study aims to evaluate the effectiveness of LLMs and traditional deep learning models in AE extraction, and to assess the impact of ensembling these models on performance. In this study, we utilized reports and posts from the VAERS (n=621), Twitter (n=9,133), and Reddit (n=131) as our corpora. Our goal was to extract three types of entities: "vaccine", "shot", and "ae". We explored and fine-tuned (except GPT-4) multiple LLMs, including GPT-2, GPT-3.5, GPT-4, and Llama-2, as well as traditional deep learning models like RNN and BioBERT. To enhance performance, we created ensembles of the three models with the best performance. For evaluation, we used strict and relaxed F1 scores to evaluate the performance for each entity type, and micro-average F1 was used to assess the overall performance. The ensemble model achieved the highest performance in "vaccine", "shot", and "ae" with strict F1-scores of 0.878, 0.930, and 0.925, respectively, along with a micro-average score of 0.903. In conclusion, this study demonstrates the effectiveness and robustness of ensembling fine-tuned traditional deep learning models and LLMs, for extracting AE-related information. This study contributes to the advancement of biomedical natural language processing, providing valuable insights into improving AE extraction from text data for pharmacovigilance and public health surveillance.
Relation Extraction Using Large Language Models: A Case Study on Acupuncture Point Locations
Apr 15, 2024Abstract:In acupuncture therapy, the accurate location of acupoints is essential for its effectiveness. The advanced language understanding capabilities of large language models (LLMs) like Generative Pre-trained Transformers (GPT) present a significant opportunity for extracting relations related to acupoint locations from textual knowledge sources. This study aims to compare the performance of GPT with traditional deep learning models (Long Short-Term Memory (LSTM) and Bidirectional Encoder Representations from Transformers for Biomedical Text Mining (BioBERT)) in extracting acupoint-related location relations and assess the impact of pretraining and fine-tuning on GPT's performance. We utilized the World Health Organization Standard Acupuncture Point Locations in the Western Pacific Region (WHO Standard) as our corpus, which consists of descriptions of 361 acupoints. Five types of relations ('direction_of,' 'distance_of,' 'part_of,' 'near_acupoint,' and 'located_near') (n= 3,174) between acupoints were annotated. Five models were compared: BioBERT, LSTM, pre-trained GPT-3.5, fine-tuned GPT-3.5, as well as pre-trained GPT-4. Performance metrics included micro-average exact match precision, recall, and F1 scores. Our results demonstrate that fine-tuned GPT-3.5 consistently outperformed other models in F1 scores across all relation types. Overall, it achieved the highest micro-average F1 score of 0.92. This study underscores the effectiveness of LLMs like GPT in extracting relations related to acupoint locations, with implications for accurately modeling acupuncture knowledge and promoting standard implementation in acupuncture training and practice. The findings also contribute to advancing informatics applications in traditional and complementary medicine, showcasing the potential of LLMs in natural language processing.
Uncovering Misattributed Suicide Causes through Annotation Inconsistency Detection in Death Investigation Notes
Mar 29, 2024



Abstract:Data accuracy is essential for scientific research and policy development. The National Violent Death Reporting System (NVDRS) data is widely used for discovering the patterns and causes of death. Recent studies suggested the annotation inconsistencies within the NVDRS and the potential impact on erroneous suicide-cause attributions. We present an empirical Natural Language Processing (NLP) approach to detect annotation inconsistencies and adopt a cross-validation-like paradigm to identify problematic instances. We analyzed 267,804 suicide death incidents between 2003 and 2020 from the NVDRS. Our results showed that incorporating the target state's data into training the suicide-crisis classifier brought an increase of 5.4% to the F-1 score on the target state's test set and a decrease of 1.1% on other states' test set. To conclude, we demonstrated the annotation inconsistencies in NVDRS's death investigation notes, identified problematic instances, evaluated the effectiveness of correcting problematic instances, and eventually proposed an NLP improvement solution.
AE-GPT: Using Large Language Models to Extract Adverse Events from Surveillance Reports-A Use Case with Influenza Vaccine Adverse Events
Sep 28, 2023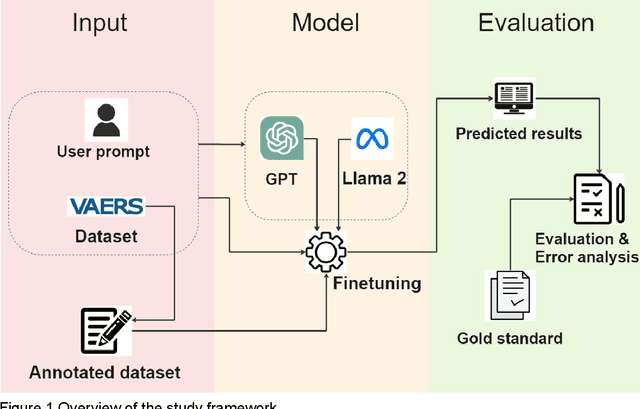
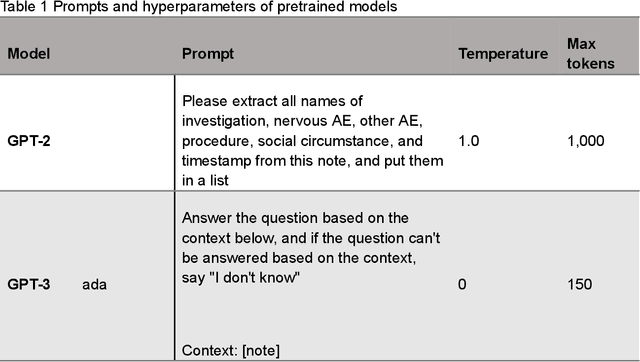


Abstract:Though Vaccines are instrumental in global health, mitigating infectious diseases and pandemic outbreaks, they can occasionally lead to adverse events (AEs). Recently, Large Language Models (LLMs) have shown promise in effectively identifying and cataloging AEs within clinical reports. Utilizing data from the Vaccine Adverse Event Reporting System (VAERS) from 1990 to 2016, this study particularly focuses on AEs to evaluate LLMs' capability for AE extraction. A variety of prevalent LLMs, including GPT-2, GPT-3 variants, GPT-4, and Llama 2, were evaluated using Influenza vaccine as a use case. The fine-tuned GPT 3.5 model (AE-GPT) stood out with a 0.704 averaged micro F1 score for strict match and 0.816 for relaxed match. The encouraging performance of the AE-GPT underscores LLMs' potential in processing medical data, indicating a significant stride towards advanced AE detection, thus presumably generalizable to other AE extraction tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge