Evan Yu
Prompting Large Language Models for Clinical Temporal Relation Extraction
Dec 04, 2024Abstract:Objective: This paper aims to prompt large language models (LLMs) for clinical temporal relation extraction (CTRE) in both few-shot and fully supervised settings. Materials and Methods: This study utilizes four LLMs: Encoder-based GatorTron-Base (345M)/Large (8.9B); Decoder-based LLaMA3-8B/MeLLaMA-13B. We developed full (FFT) and parameter-efficient (PEFT) fine-tuning strategies and evaluated these strategies on the 2012 i2b2 CTRE task. We explored four fine-tuning strategies for GatorTron-Base: (1) Standard Fine-Tuning, (2) Hard-Prompting with Unfrozen LLMs, (3) Soft-Prompting with Frozen LLMs, and (4) Low-Rank Adaptation (LoRA) with Frozen LLMs. For GatorTron-Large, we assessed two PEFT strategies-Soft-Prompting and LoRA with Frozen LLMs-leveraging Quantization techniques. Additionally, LLaMA3-8B and MeLLaMA-13B employed two PEFT strategies: LoRA strategy with Quantization (QLoRA) applied to Frozen LLMs using instruction tuning and standard fine-tuning. Results: Under fully supervised settings, Hard-Prompting with Unfrozen GatorTron-Base achieved the highest F1 score (89.54%), surpassing the SOTA model (85.70%) by 3.74%. Additionally, two variants of QLoRA adapted to GatorTron-Large and Standard Fine-Tuning of GatorTron-Base exceeded the SOTA model by 2.36%, 1.88%, and 0.25%, respectively. Decoder-based models with frozen parameters outperformed their Encoder-based counterparts in this setting; however, the trend reversed in few-shot scenarios. Discussions and Conclusions: This study presented new methods that significantly improved CTRE performance, benefiting downstream tasks reliant on CTRE systems. The findings underscore the importance of selecting appropriate models and fine-tuning strategies based on task requirements and data availability. Future work will explore larger models and broader CTRE applications.
Explainable Graph Neural Network for Alzheimer's Disease And Related Dementias Risk Prediction
Sep 18, 2023Abstract:Alzheimer's disease and related dementias (ADRD) ranks as the sixth leading cause of death in the US, underlining the importance of accurate ADRD risk prediction. While recent advancement in ADRD risk prediction have primarily relied on imaging analysis, yet not all patients undergo medical imaging before an ADRD diagnosis. Merging machine learning with claims data can reveal additional risk factors and uncover interconnections among diverse medical codes. Our goal is to utilize Graph Neural Networks (GNNs) with claims data for ADRD risk prediction. Addressing the lack of human-interpretable reasons behind these predictions, we introduce an innovative method to evaluate relationship importance and its influence on ADRD risk prediction, ensuring comprehensive interpretation. We employed Variationally Regularized Encoder-decoder Graph Neural Network (VGNN) for estimating ADRD likelihood. We created three scenarios to assess the model's efficiency, using Random Forest and Light Gradient Boost Machine as baselines. We further used our relation importance method to clarify the key relationships for ADRD risk prediction. VGNN surpassed other baseline models by 10% in the area under the receiver operating characteristic. The integration of the GNN model and relation importance interpretation could potentially play an essential role in providing valuable insight into factors that may contribute to or delay ADRD progression. Employing a GNN approach with claims data enhances ADRD risk prediction and provides insights into the impact of interconnected medical code relationships. This methodology not only enables ADRD risk modeling but also shows potential for other image analysis predictions using claims data.
Unsupervised deep learning for Bayesian brain MRI segmentation
Apr 25, 2019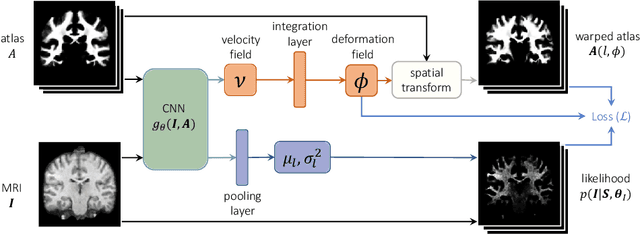

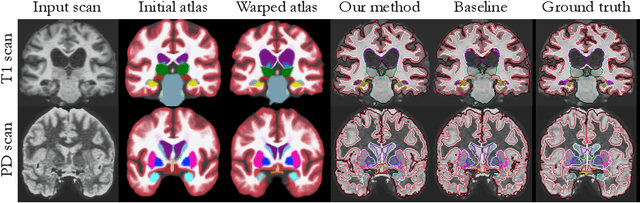
Abstract:Probabilistic atlas priors have been commonly used to derive adaptive and robust brain MRI segmentation algorithms. Widely-used neuroimage analysis pipelines rely heavily on these techniques, which are often computationally expensive. In contrast, there has been a recent surge of approaches that leverage deep learning to implement segmentation tools that are computationally efficient at test time. However, most of these strategies rely on learning from manually annotated images. These supervised deep learning methods are therefore sensitive to the intensity profiles in the training dataset. To develop a deep learning-based segmentation model for a new image dataset (e.g., of different contrast), one usually needs to create a new labeled training dataset, which can be prohibitively expensive, or rely on suboptimal ad hoc adaptation or augmentation approaches. In this paper, we propose an alternative strategy that combines a conventional probabilistic atlas-based segmentation with deep learning, enabling one to train a segmentation model for new MRI scans without the need for any manually segmented images. Our experiments include thousands of brain MRI scans and demonstrate that the proposed method achieves good accuracy for a brain MRI segmentation task for different MRI contrasts, requiring only approximately 15 seconds at test time on a GPU. The code is freely available at http://voxelmorph.mit.edu.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge