Juan Eugenio Iglesias
Centre for Medical Image Computing, Department of Computer Science, University College London, UK, Martinos Center for Biomedical Imaging, Massachusetts General Hospital and Harvard Medical School, Boston, USA, Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Boston, USA
Reference-Free 3D Reconstruction of Brain Dissection Photographs with Machine Learning
Mar 13, 2025Abstract:Correlation of neuropathology with MRI has the potential to transfer microscopic signatures of pathology to invivo scans. Recently, a classical registration method has been proposed, to build these correlations from 3D reconstructed stacks of dissection photographs, which are routinely taken at brain banks. These photographs bypass the need for exvivo MRI, which is not widely accessible. However, this method requires a full stack of brain slabs and a reference mask (e.g., acquired with a surface scanner), which severely limits the applicability of the technique. Here we propose RefFree, a dissection photograph reconstruction method without external reference. RefFree is a learning approach that estimates the 3D coordinates in the atlas space for every pixel in every photograph; simple least-squares fitting can then be used to compute the 3D reconstruction. As a by-product, RefFree also produces an atlas-based segmentation of the reconstructed stack. RefFree is trained on synthetic photographs generated from digitally sliced 3D MRI data, with randomized appearance for enhanced generalization ability. Experiments on simulated and real data show that RefFree achieves performance comparable to the baseline method without an explicit reference while also enabling reconstruction of partial stacks. Our code is available at https://github.com/lintian-a/reffree.
Learn2Synth: Learning Optimal Data Synthesis Using Hypergradients
Nov 23, 2024



Abstract:Domain randomization through synthesis is a powerful strategy to train networks that are unbiased with respect to the domain of the input images. Randomization allows networks to see a virtually infinite range of intensities and artifacts during training, thereby minimizing overfitting to appearance and maximizing generalization to unseen data. While powerful, this approach relies on the accurate tuning of a large set of hyper-parameters governing the probabilistic distribution of the synthesized images. Instead of manually tuning these parameters, we introduce Learn2Synth, a novel procedure in which synthesis parameters are learned using a small set of real labeled data. Unlike methods that impose constraints to align synthetic data with real data (e.g., contrastive or adversarial techniques), which risk misaligning the image and its label map, we tune an augmentation engine such that a segmentation network trained on synthetic data has optimal accuracy when applied to real data. This approach allows the training procedure to benefit from real labeled examples, without ever using these real examples to train the segmentation network, which avoids biasing the network towards the properties of the training set. Specifically, we develop both parametric and nonparametric strategies to augment the synthetic images, enhancing the segmentation network's performance. Experimental results on both synthetic and real-world datasets demonstrate the effectiveness of this learning strategy. Code is available at: https://github.com/HuXiaoling/Learn2Synth.
A Constrast-Agnostic Method for Ultra-High Resolution Claustrum Segmentation
Nov 23, 2024



Abstract:The claustrum is a band-like gray matter structure located between putamen and insula whose exact functions are still actively researched. Its sheet-like structure makes it barely visible in in vivo Magnetic Resonance Imaging (MRI) scans at typical resolutions and neuroimaging tools for its study, including methods for automatic segmentation, are currently very limited. In this paper, we propose a contrast- and resolution-agnostic method for claustrum segmentation at ultra-high resolution (0.35 mm isotropic); the method is based on the SynthSeg segmentation framework (Billot et al., 2023), which leverages the use of synthetic training intensity images to achieve excellent generalization. In particular, SynthSeg requires only label maps to be trained, since corresponding intensity images are synthesized on the fly with random contrast and resolution. We trained a deep learning network for automatic claustrum segmentation, using claustrum manual labels obtained from 18 ultra-high resolution MRI scans (mostly ex vivo). We demonstrated the method to work on these 18 high resolution cases (Dice score = 0.632, mean surface distance = 0.458 mm, and volumetric similarity = 0.867 using 6-fold Cross Validation (CV)), and also on in vivo T1-weighted MRI scans at typical resolutions (~1 mm isotropic). We also demonstrated that the method is robust in a test-retest setting and when applied to multimodal imaging (T2-weighted, Proton Density and quantitative T1 scans). To the best of our knowledge this is the first accurate method for automatic ultra-high resolution claustrum segmentation, which is robust against changes in contrast and resolution. The method is released at https://github.com/chiara-mauri/claustrum_segmentation and as part of the neuroimaging package Freesurfer (Fischl, 2012).
TV-based Deep 3D Self Super-Resolution for fMRI
Oct 05, 2024Abstract:While functional Magnetic Resonance Imaging (fMRI) offers valuable insights into cognitive processes, its inherent spatial limitations pose challenges for detailed analysis of the fine-grained functional architecture of the brain. More specifically, MRI scanner and sequence specifications impose a trade-off between temporal resolution, spatial resolution, signal-to-noise ratio, and scan time. Deep Learning (DL) Super-Resolution (SR) methods have emerged as a promising solution to enhance fMRI resolution, generating high-resolution (HR) images from low-resolution (LR) images typically acquired with lower scanning times. However, most existing SR approaches depend on supervised DL techniques, which require training ground truth (GT) HR data, which is often difficult to acquire and simultaneously sets a bound for how far SR can go. In this paper, we introduce a novel self-supervised DL SR model that combines a DL network with an analytical approach and Total Variation (TV) regularization. Our method eliminates the need for external GT images, achieving competitive performance compared to supervised DL techniques and preserving the functional maps.
Recon-all-clinical: Cortical surface reconstruction and analysis of heterogeneous clinical brain MRI
Sep 05, 2024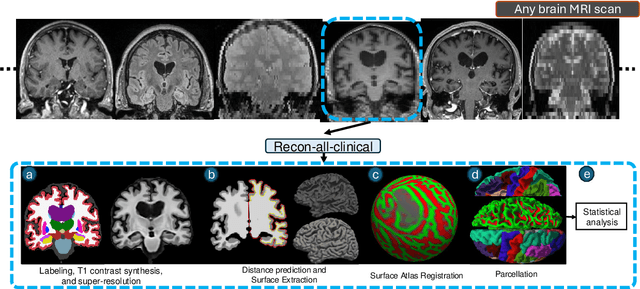
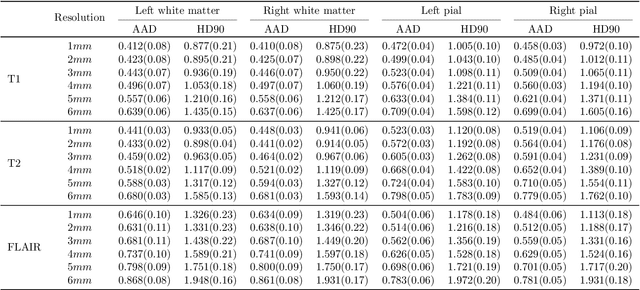
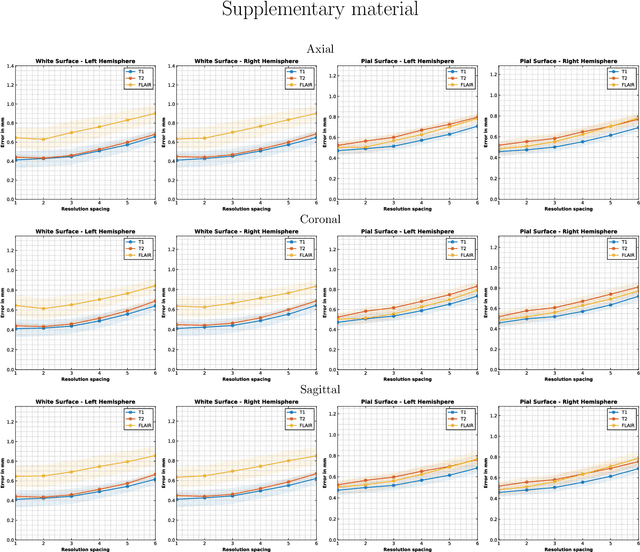
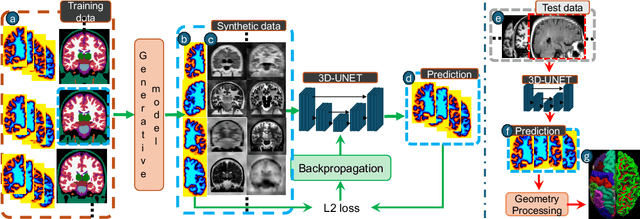
Abstract:Surface-based analysis of the cerebral cortex is ubiquitous in human neuroimaging with MRI. It is crucial for cortical registration, parcellation, and thickness estimation. Traditionally, these analyses require high-resolution, isotropic scans with good gray-white matter contrast, typically a 1mm T1-weighted scan. This excludes most clinical MRI scans, which are often anisotropic and lack the necessary T1 contrast. To enable large-scale neuroimaging studies using vast clinical data, we introduce recon-all-clinical, a novel method for cortical reconstruction, registration, parcellation, and thickness estimation in brain MRI scans of any resolution and contrast. Our approach employs a hybrid analysis method that combines a convolutional neural network (CNN) trained with domain randomization to predict signed distance functions (SDFs) and classical geometry processing for accurate surface placement while maintaining topological and geometric constraints. The method does not require retraining for different acquisitions, thus simplifying the analysis of heterogeneous clinical datasets. We tested recon-all-clinical on multiple datasets, including over 19,000 clinical scans. The method consistently produced precise cortical reconstructions and high parcellation accuracy across varied MRI contrasts and resolutions. Cortical thickness estimates are precise enough to capture aging effects independently of MRI contrast, although accuracy varies with slice thickness. Our method is publicly available at https://surfer.nmr.mgh.harvard.edu/fswiki/recon-all-clinical, enabling researchers to perform detailed cortical analysis on the huge amounts of already existing clinical MRI scans. This advancement may be particularly valuable for studying rare diseases and underrepresented populations where research-grade MRI data is scarce.
BraTS-PEDs: Results of the Multi-Consortium International Pediatric Brain Tumor Segmentation Challenge 2023
Jul 11, 2024

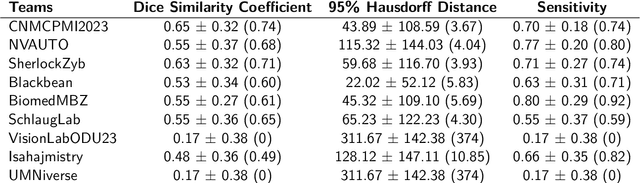
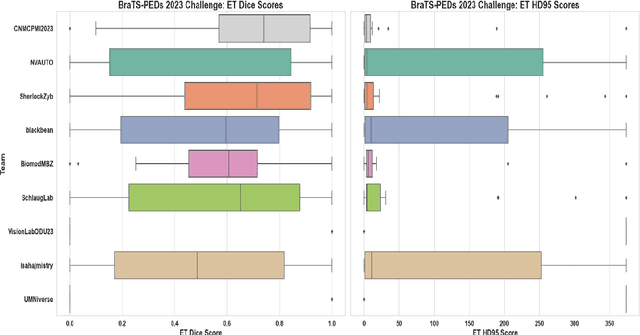
Abstract:Pediatric central nervous system tumors are the leading cause of cancer-related deaths in children. The five-year survival rate for high-grade glioma in children is less than 20%. The development of new treatments is dependent upon multi-institutional collaborative clinical trials requiring reproducible and accurate centralized response assessment. We present the results of the BraTS-PEDs 2023 challenge, the first Brain Tumor Segmentation (BraTS) challenge focused on pediatric brain tumors. This challenge utilized data acquired from multiple international consortia dedicated to pediatric neuro-oncology and clinical trials. BraTS-PEDs 2023 aimed to evaluate volumetric segmentation algorithms for pediatric brain gliomas from magnetic resonance imaging using standardized quantitative performance evaluation metrics employed across the BraTS 2023 challenges. The top-performing AI approaches for pediatric tumor analysis included ensembles of nnU-Net and Swin UNETR, Auto3DSeg, or nnU-Net with a self-supervised framework. The BraTSPEDs 2023 challenge fostered collaboration between clinicians (neuro-oncologists, neuroradiologists) and AI/imaging scientists, promoting faster data sharing and the development of automated volumetric analysis techniques. These advancements could significantly benefit clinical trials and improve the care of children with brain tumors.
High-resolution segmentations of the hypothalamus and its subregions for training of segmentation models
Jun 27, 2024Abstract:Segmentation of brain structures on magnetic resonance imaging (MRI) is a highly relevant neuroimaging topic, as it is a prerequisite for different analyses such as volumetry or shape analysis. Automated segmentation facilitates the study of brain structures in larger cohorts when compared with manual segmentation, which is time-consuming. However, the development of most automated methods relies on large and manually annotated datasets, which limits the generalizability of these methods. Recently, new techniques using synthetic images have emerged, reducing the need for manual annotation. Here we provide HELM, Hypothalamic ex vivo Label Maps, a dataset composed of label maps built from publicly available ultra-high resolution ex vivo MRI from 10 whole hemispheres, which can be used to develop segmentation methods using synthetic data. The label maps are obtained with a combination of manual labels for the hypothalamic regions and automated segmentations for the rest of the brain, and mirrored to simulate entire brains. We also provide the pre-processed ex vivo scans, as this dataset can support future projects to include other structures after these are manually segmented.
Probabilistic Contrastive Learning with Explicit Concentration on the Hypersphere
May 26, 2024Abstract:Self-supervised contrastive learning has predominantly adopted deterministic methods, which are not suited for environments characterized by uncertainty and noise. This paper introduces a new perspective on incorporating uncertainty into contrastive learning by embedding representations within a spherical space, inspired by the von Mises-Fisher distribution (vMF). We introduce an unnormalized form of vMF and leverage the concentration parameter, kappa, as a direct, interpretable measure to quantify uncertainty explicitly. This approach not only provides a probabilistic interpretation of the embedding space but also offers a method to calibrate model confidence against varying levels of data corruption and characteristics. Our empirical results demonstrate that the estimated concentration parameter correlates strongly with the degree of unforeseen data corruption encountered at test time, enables failure analysis, and enhances existing out-of-distribution detection methods.
Analysis of the BraTS 2023 Intracranial Meningioma Segmentation Challenge
May 16, 2024



Abstract:We describe the design and results from the BraTS 2023 Intracranial Meningioma Segmentation Challenge. The BraTS Meningioma Challenge differed from prior BraTS Glioma challenges in that it focused on meningiomas, which are typically benign extra-axial tumors with diverse radiologic and anatomical presentation and a propensity for multiplicity. Nine participating teams each developed deep-learning automated segmentation models using image data from the largest multi-institutional systematically expert annotated multilabel multi-sequence meningioma MRI dataset to date, which included 1000 training set cases, 141 validation set cases, and 283 hidden test set cases. Each case included T2, T2/FLAIR, T1, and T1Gd brain MRI sequences with associated tumor compartment labels delineating enhancing tumor, non-enhancing tumor, and surrounding non-enhancing T2/FLAIR hyperintensity. Participant automated segmentation models were evaluated and ranked based on a scoring system evaluating lesion-wise metrics including dice similarity coefficient (DSC) and 95% Hausdorff Distance. The top ranked team had a lesion-wise median dice similarity coefficient (DSC) of 0.976, 0.976, and 0.964 for enhancing tumor, tumor core, and whole tumor, respectively and a corresponding average DSC of 0.899, 0.904, and 0.871, respectively. These results serve as state-of-the-art benchmarks for future pre-operative meningioma automated segmentation algorithms. Additionally, we found that 1286 of 1424 cases (90.3%) had at least 1 compartment voxel abutting the edge of the skull-stripped image edge, which requires further investigation into optimal pre-processing face anonymization steps.
The Brain Tumor Segmentation in Pediatrics (BraTS-PEDs) Challenge: Focus on Pediatrics (CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs)
Apr 29, 2024Abstract:Pediatric tumors of the central nervous system are the most common cause of cancer-related death in children. The five-year survival rate for high-grade gliomas in children is less than 20%. Due to their rarity, the diagnosis of these entities is often delayed, their treatment is mainly based on historic treatment concepts, and clinical trials require multi-institutional collaborations. Here we present the CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs challenge, focused on pediatric brain tumors with data acquired across multiple international consortia dedicated to pediatric neuro-oncology and clinical trials. The CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs challenge brings together clinicians and AI/imaging scientists to lead to faster development of automated segmentation techniques that could benefit clinical trials, and ultimately the care of children with brain tumors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge