Yael Balbastre
Learn2Synth: Learning Optimal Data Synthesis Using Hypergradients
Nov 23, 2024



Abstract:Domain randomization through synthesis is a powerful strategy to train networks that are unbiased with respect to the domain of the input images. Randomization allows networks to see a virtually infinite range of intensities and artifacts during training, thereby minimizing overfitting to appearance and maximizing generalization to unseen data. While powerful, this approach relies on the accurate tuning of a large set of hyper-parameters governing the probabilistic distribution of the synthesized images. Instead of manually tuning these parameters, we introduce Learn2Synth, a novel procedure in which synthesis parameters are learned using a small set of real labeled data. Unlike methods that impose constraints to align synthetic data with real data (e.g., contrastive or adversarial techniques), which risk misaligning the image and its label map, we tune an augmentation engine such that a segmentation network trained on synthetic data has optimal accuracy when applied to real data. This approach allows the training procedure to benefit from real labeled examples, without ever using these real examples to train the segmentation network, which avoids biasing the network towards the properties of the training set. Specifically, we develop both parametric and nonparametric strategies to augment the synthetic images, enhancing the segmentation network's performance. Experimental results on both synthetic and real-world datasets demonstrate the effectiveness of this learning strategy. Code is available at: https://github.com/HuXiaoling/Learn2Synth.
A label-free and data-free training strategy for vasculature segmentation in serial sectioning OCT data
May 22, 2024Abstract:Serial sectioning Optical Coherence Tomography (sOCT) is a high-throughput, label free microscopic imaging technique that is becoming increasingly popular to study post-mortem neurovasculature. Quantitative analysis of the vasculature requires highly accurate segmentation; however, sOCT has low signal-to-noise-ratio and displays a wide range of contrasts and artifacts that depend on acquisition parameters. Furthermore, labeled data is scarce and extremely time consuming to generate. Here, we leverage synthetic datasets of vessels to train a deep learning segmentation model. We construct the vessels with semi-realistic splines that simulate the vascular geometry and compare our model with realistic vascular labels generated by constrained constructive optimization. Both approaches yield similar Dice scores, although with very different false positive and false negative rates. This method addresses the complexity inherent in OCT images and paves the way for more accurate and efficient analysis of neurovascular structures.
Fitting Segmentation Networks on Varying Image Resolutions using Splatting
Jun 15, 2022


Abstract:Data used in image segmentation are not always defined on the same grid. This is particularly true for medical images, where the resolution, field-of-view and orientation can differ across channels and subjects. Images and labels are therefore commonly resampled onto the same grid, as a pre-processing step. However, the resampling operation introduces partial volume effects and blurring, thereby changing the effective resolution and reducing the contrast between structures. In this paper we propose a splat layer, which automatically handles resolution mismatches in the input data. This layer pushes each image onto a mean space where the forward pass is performed. As the splat operator is the adjoint to the resampling operator, the mean-space prediction can be pulled back to the native label space, where the loss function is computed. Thus, the need for explicit resolution adjustment using interpolation is removed. We show on two publicly available datasets, with simulated and real multi-modal magnetic resonance images, that this model improves segmentation results compared to resampling as a pre-processing step.
Factorisation-based Image Labelling
Nov 19, 2021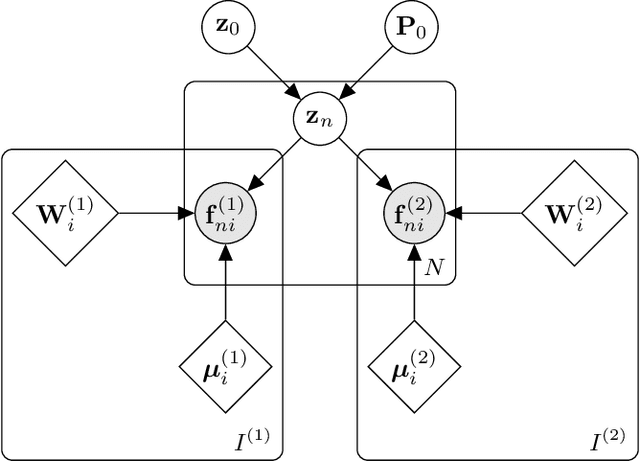
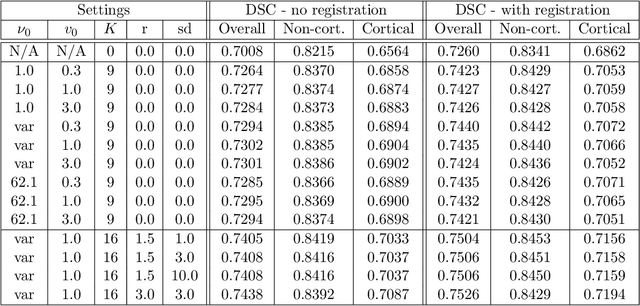
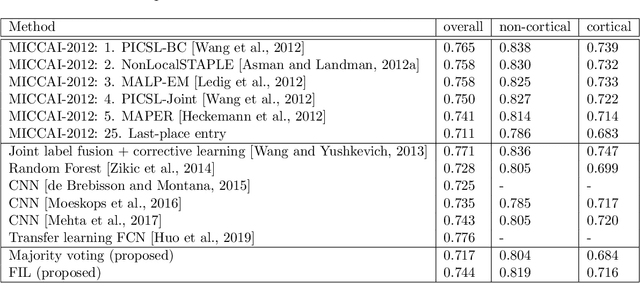
Abstract:Segmentation of brain magnetic resonance images (MRI) into anatomical regions is a useful task in neuroimaging. Manual annotation is time consuming and expensive, so having a fully automated and general purpose brain segmentation algorithm is highly desirable. To this end, we propose a patched-based label propagation approach based on a generative model with latent variables. Once trained, our Factorisation-based Image Labelling (FIL) model is able to label target images with a variety of image contrasts. We compare the effectiveness of our proposed model against the state-of-the-art using data from the MICCAI 2012 Grand Challenge and Workshop on Multi-Atlas Labeling. As our approach is intended to be general purpose, we also assess how well it can handle domain shift by labelling images of the same subjects acquired with different MR contrasts.
Model-based multi-parameter mapping
Feb 02, 2021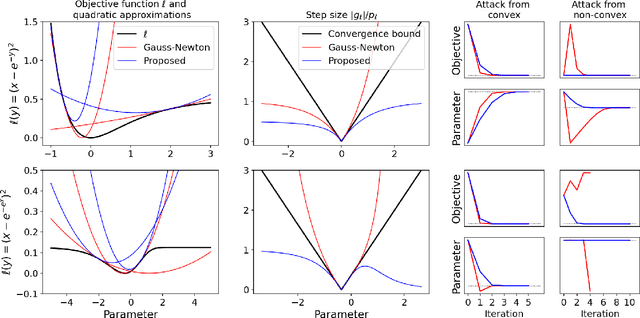
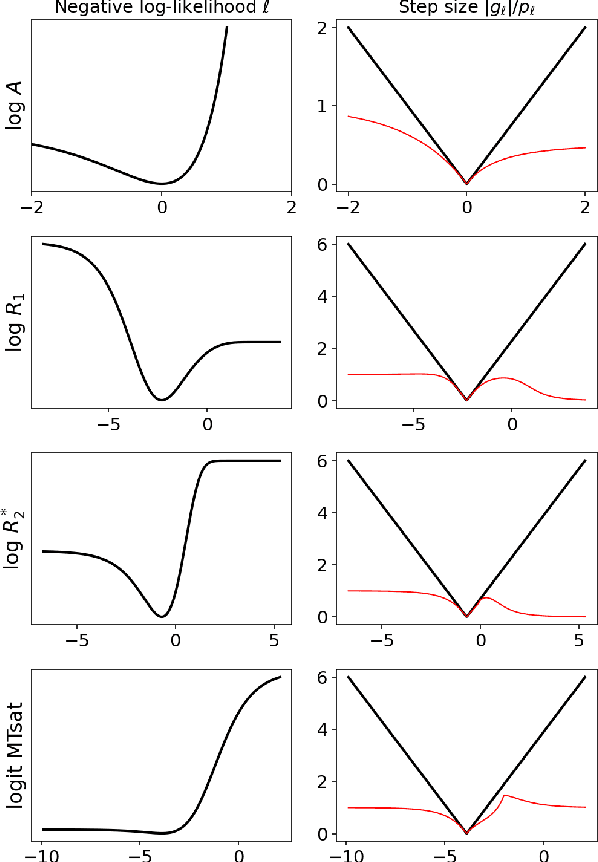
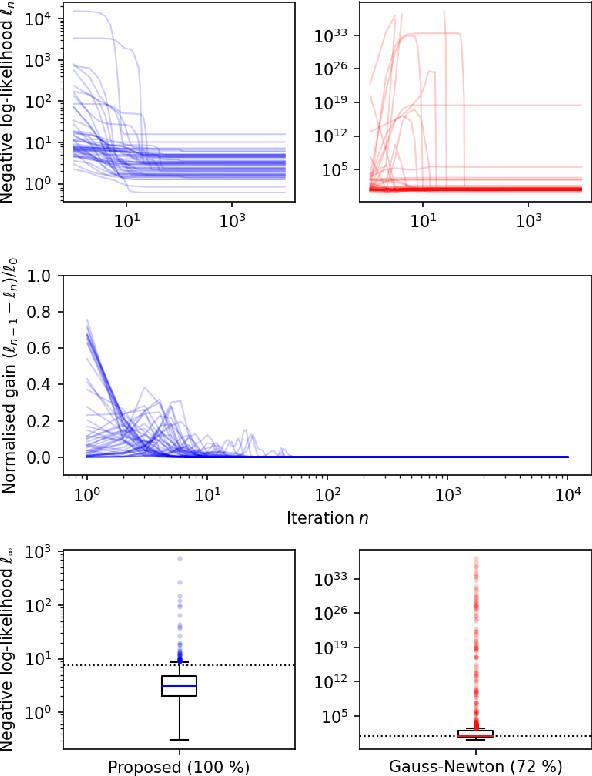
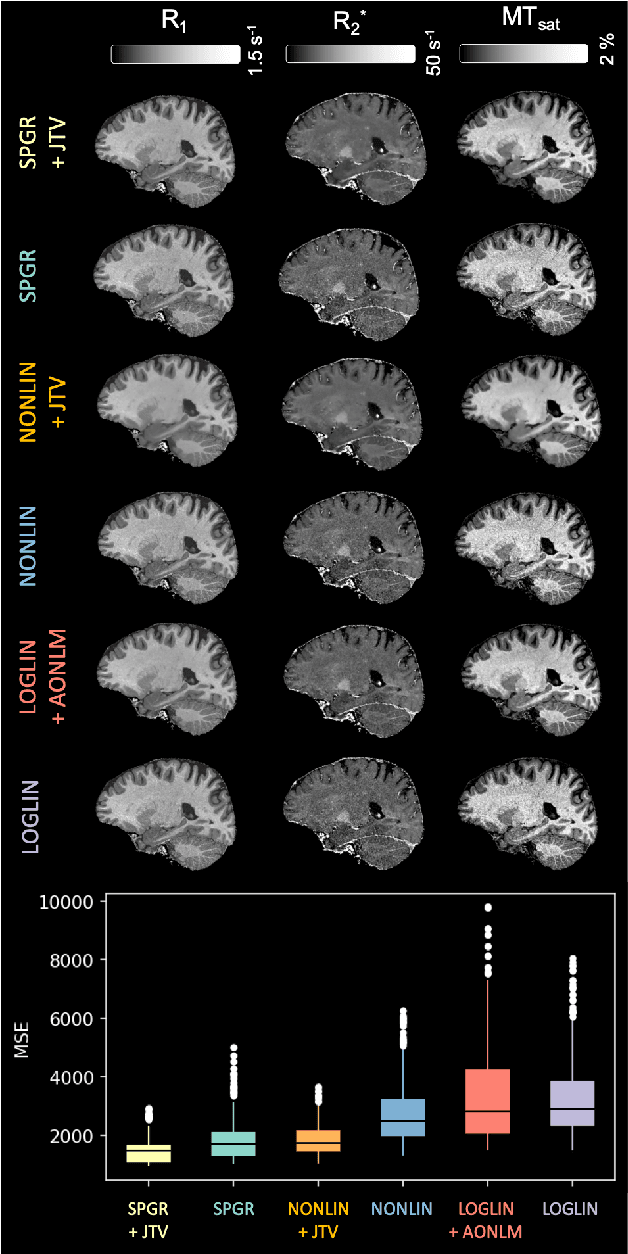
Abstract:Quantitative MR imaging is increasingly favoured for its richer information content and standardised measures. However, extracting quantitative parameters such as the longitudinal relaxation rate (R1), apparent transverse relaxation rate (R2*), or magnetisation-transfer saturation (MTsat) involves inverting a highly non-linear function. Estimations often assume noise-free measurements and use subsets of the data to solve for different quantities in isolation, with error propagating through each computation. Instead, a probabilistic generative model of the entire dataset can be formulated and inverted to jointly recover parameter estimates with a well-defined probabilistic meaning (e.g., maximum likelihood or maximum a posteriori). In practice, iterative methods must be used but convergence is difficult due to the non-convexity of the log-likelihood; yet, we show that it can be achieved thanks to a novel approximate Hessian and, with it, reliable parameter estimates obtained. Here, we demonstrate the utility of this flexible framework in the context of the popular multi-parameter mapping framework and further show how to incorporate a denoising prior and predict posterior uncertainty. Our implementation uses a PyTorch backend and benefits from GPU acceleration. It is available at https://github.com/balbasty/nitorch.
Joint super-resolution and synthesis of 1 mm isotropic MP-RAGE volumes from clinical MRI exams with scans of different orientation, resolution and contrast
Dec 24, 2020



Abstract:Most existing algorithms for automatic 3D morphometry of human brain MRI scans are designed for data with near-isotropic voxels at approximately 1 mm resolution, and frequently have contrast constraints as well - typically requiring T1 scans (e.g., MP-RAGE). This limitation prevents the analysis of millions of MRI scans acquired with large inter-slice spacing ("thick slice") in clinical settings every year. The inability to quantitatively analyze these scans hinders the adoption of quantitative neuroimaging in healthcare, and precludes research studies that could attain huge sample sizes and hence greatly improve our understanding of the human brain. Recent advances in CNNs are producing outstanding results in super-resolution and contrast synthesis of MRI. However, these approaches are very sensitive to the contrast, resolution and orientation of the input images, and thus do not generalize to diverse clinical acquisition protocols - even within sites. Here we present SynthSR, a method to train a CNN that receives one or more thick-slice scans with different contrast, resolution and orientation, and produces an isotropic scan of canonical contrast (typically a 1 mm MP-RAGE). The presented method does not require any preprocessing, e.g., skull stripping or bias field correction. Crucially, SynthSR trains on synthetic input images generated from 3D segmentations, and can thus be used to train CNNs for any combination of contrasts, resolutions and orientations without high-resolution training data. We test the images generated with SynthSR in an array of common downstream analyses, and show that they can be reliably used for subcortical segmentation and volumetry, image registration (e.g., for tensor-based morphometry), and, if some image quality requirements are met, even cortical thickness morphometry. The source code is publicly available at github.com/BBillot/SynthSR.
A Tool for Super-Resolving Multimodal Clinical MRI
Sep 03, 2019



Abstract:We present a tool for resolution recovery in multimodal clinical magnetic resonance imaging (MRI). Such images exhibit great variability, both biological and instrumental. This variability makes automated processing with neuroimaging analysis software very challenging. This leaves intelligence extractable only from large-scale analyses of clinical data untapped, and impedes the introduction of automated predictive systems in clinical care. The tool presented in this paper enables such processing, via inference in a generative model of thick-sliced, multi-contrast MR scans. All model parameters are estimated from the observed data, without the need for manual tuning. The model-driven nature of the approach means that no type of training is needed for applicability to the diversity of MR contrasts present in a clinical context. We show on simulated data that the proposed approach outperforms conventional model-based techniques, and on a large hospital dataset of multimodal MRIs that the tool can successfully super-resolve very thick-sliced images. The implementation is available from https://github.com/brudfors/spm_superres.
Empirical Bayesian Mixture Models for Medical Image Translation
Aug 16, 2019



Abstract:Automatically generating one medical imaging modality from another is known as medical image translation, and has numerous interesting applications. This paper presents an interpretable generative modelling approach to medical image translation. By allowing a common model for group-wise normalisation and segmentation of brain scans to handle missing data, the model allows for predicting entirely missing modalities from one, or a few, MR contrasts. Furthermore, the model can be trained on a fairly small number of subjects. The proposed model is validated on three clinically relevant scenarios. Results appear promising and show that a principled, probabilistic model of the relationship between multi-channel signal intensities can be used to infer missing modalities -- both MR contrasts and CT images.
MRI Super-Resolution using Multi-Channel Total Variation
Oct 18, 2018



Abstract:This paper presents a generative model for super-resolution in routine clinical magnetic resonance images (MRI), of arbitrary orientation and contrast. The model recasts the recovery of high resolution images as an inverse problem, in which a forward model simulates the slice-select profile of the MR scanner. The paper introduces a prior based on multi-channel total variation for MRI super-resolution. Bias-variance trade-off is handled by estimating hyper-parameters from the low resolution input scans. The model was validated on a large database of brain images. The validation showed that the model can improve brain segmentation, that it can recover anatomical information between images of different MR contrasts, and that it generalises well to the large variability present in MR images of different subjects. The implementation is freely available at https://github.com/WCHN/mtv-preproc
An Algorithm for Learning Shape and Appearance Models without Annotations
Jul 27, 2018



Abstract:This paper presents a framework for automatically learning shape and appearance models for medical (and certain other) images. It is based on the idea that having a more accurate shape and appearance model leads to more accurate image registration, which in turn leads to a more accurate shape and appearance model. This leads naturally to an iterative scheme, which is based on a probabilistic generative model that is fit using Gauss-Newton updates within an EM-like framework. It was developed with the aim of enabling distributed privacy-preserving analysis of brain image data, such that shared information (shape and appearance basis functions) may be passed across sites, whereas latent variables that encode individual images remain secure within each site. These latent variables are proposed as features for privacy-preserving data mining applications. The approach is demonstrated qualitatively on the KDEF dataset of 2D face images, showing that it can align images that traditionally require shape and appearance models trained using manually annotated data (manually defined landmarks etc.). It is applied to MNIST dataset of handwritten digits to show its potential for machine learning applications, particularly when training data is limited. The model is able to handle ``missing data'', which allows it to be cross-validated according to how well it can predict left-out voxels. The suitability of the derived features for classifying individuals into patient groups was assessed by applying it to a dataset of over 1,900 segmented T1-weighted MR images, which included images from the COBRE and ABIDE datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge