Xiaoling Hu
Department of Biomedical Engineering, The Hong Kong Polytechnic University, China
Improving Neuropathological Reconstruction Fidelity via AI Slice Imputation
Jan 31, 2026Abstract:Neuropathological analyses benefit from spatially precise volumetric reconstructions that enhance anatomical delineation and improve morphometric accuracy. Our prior work has shown the feasibility of reconstructing 3D brain volumes from 2D dissection photographs. However these outputs sometimes exhibit coarse, overly smooth reconstructions of structures, especially under high anisotropy (i.e., reconstructions from thick slabs). Here, we introduce a computationally efficient super-resolution step that imputes slices to generate anatomically consistent isotropic volumes from anisotropic 3D reconstructions of dissection photographs. By training on domain-randomized synthetic data, we ensure that our method generalizes across dissection protocols and remains robust to large slab thicknesses. The imputed volumes yield improved automated segmentations, achieving higher Dice scores, particularly in cortical and white matter regions. Validation on surface reconstruction and atlas registration tasks demonstrates more accurate cortical surfaces and MRI registration. By enhancing the resolution and anatomical fidelity of photograph-based reconstructions, our approach strengthens the bridge between neuropathology and neuroimaging. Our method is publicly available at https://surfer.nmr.mgh.harvard.edu/fswiki/mri_3d_photo_recon
MATCH: Multi-faceted Adaptive Topo-Consistency for Semi-Supervised Histopathology Segmentation
Oct 02, 2025



Abstract:In semi-supervised segmentation, capturing meaningful semantic structures from unlabeled data is essential. This is particularly challenging in histopathology image analysis, where objects are densely distributed. To address this issue, we propose a semi-supervised segmentation framework designed to robustly identify and preserve relevant topological features. Our method leverages multiple perturbed predictions obtained through stochastic dropouts and temporal training snapshots, enforcing topological consistency across these varied outputs. This consistency mechanism helps distinguish biologically meaningful structures from transient and noisy artifacts. A key challenge in this process is to accurately match the corresponding topological features across the predictions in the absence of ground truth. To overcome this, we introduce a novel matching strategy that integrates spatial overlap with global structural alignment, minimizing discrepancies among predictions. Extensive experiments demonstrate that our approach effectively reduces topological errors, resulting in more robust and accurate segmentations essential for reliable downstream analysis. Code is available at \href{https://github.com/Melon-Xu/MATCH}{https://github.com/Melon-Xu/MATCH}.
Bézier Meets Diffusion: Robust Generation Across Domains for Medical Image Segmentation
Sep 26, 2025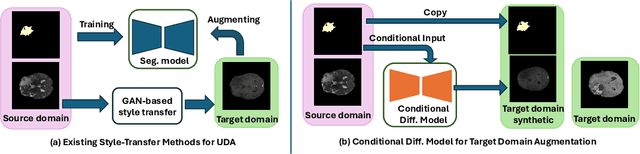
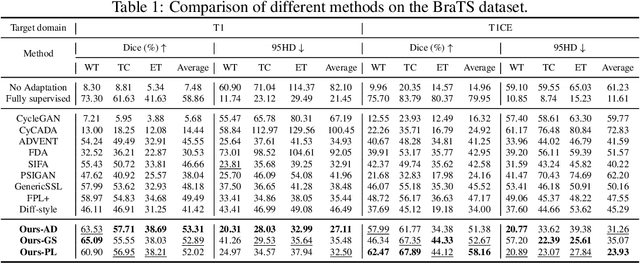
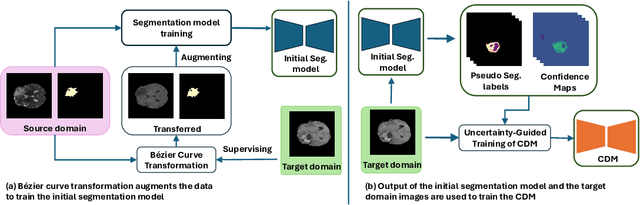
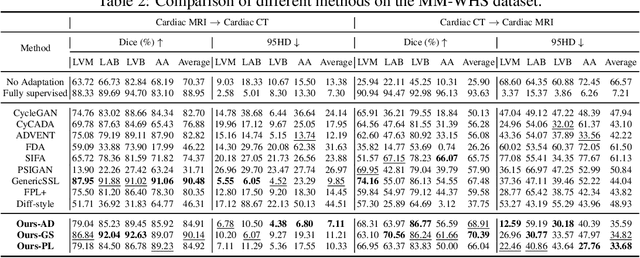
Abstract:Training robust learning algorithms across different medical imaging modalities is challenging due to the large domain gap. Unsupervised domain adaptation (UDA) mitigates this problem by using annotated images from the source domain and unlabeled images from the target domain to train the deep models. Existing approaches often rely on GAN-based style transfer, but these methods struggle to capture cross-domain mappings in regions with high variability. In this paper, we propose a unified framework, B\'ezier Meets Diffusion, for cross-domain image generation. First, we introduce a B\'ezier-curve-based style transfer strategy that effectively reduces the domain gap between source and target domains. The transferred source images enable the training of a more robust segmentation model across domains. Thereafter, using pseudo-labels generated by this segmentation model on the target domain, we train a conditional diffusion model (CDM) to synthesize high-quality, labeled target-domain images. To mitigate the impact of noisy pseudo-labels, we further develop an uncertainty-guided score matching method that improves the robustness of CDM training. Extensive experiments on public datasets demonstrate that our approach generates realistic labeled images, significantly augmenting the target domain and improving segmentation performance.
Scalable Segmentation for Ultra-High-Resolution Brain MR Images
May 27, 2025Abstract:Although deep learning has shown great success in 3D brain MRI segmentation, achieving accurate and efficient segmentation of ultra-high-resolution brain images remains challenging due to the lack of labeled training data for fine-scale anatomical structures and high computational demands. In this work, we propose a novel framework that leverages easily accessible, low-resolution coarse labels as spatial references and guidance, without incurring additional annotation cost. Instead of directly predicting discrete segmentation maps, our approach regresses per-class signed distance transform maps, enabling smooth, boundary-aware supervision. Furthermore, to enhance scalability, generalizability, and efficiency, we introduce a scalable class-conditional segmentation strategy, where the model learns to segment one class at a time conditioned on a class-specific input. This novel design not only reduces memory consumption during both training and testing, but also allows the model to generalize to unseen anatomical classes. We validate our method through comprehensive experiments on both synthetic and real-world datasets, demonstrating its superior performance and scalability compared to conventional segmentation approaches.
Biomedical Foundation Model: A Survey
Mar 03, 2025Abstract:Foundation models, first introduced in 2021, are large-scale pre-trained models (e.g., large language models (LLMs) and vision-language models (VLMs)) that learn from extensive unlabeled datasets through unsupervised methods, enabling them to excel in diverse downstream tasks. These models, like GPT, can be adapted to various applications such as question answering and visual understanding, outperforming task-specific AI models and earning their name due to broad applicability across fields. The development of biomedical foundation models marks a significant milestone in leveraging artificial intelligence (AI) to understand complex biological phenomena and advance medical research and practice. This survey explores the potential of foundation models across diverse domains within biomedical fields, including computational biology, drug discovery and development, clinical informatics, medical imaging, and public health. The purpose of this survey is to inspire ongoing research in the application of foundation models to health science.
Poststroke rehabilitative mechanisms in individualized fatigue level-controlled treadmill training -- a Rat Model Study
Feb 20, 2025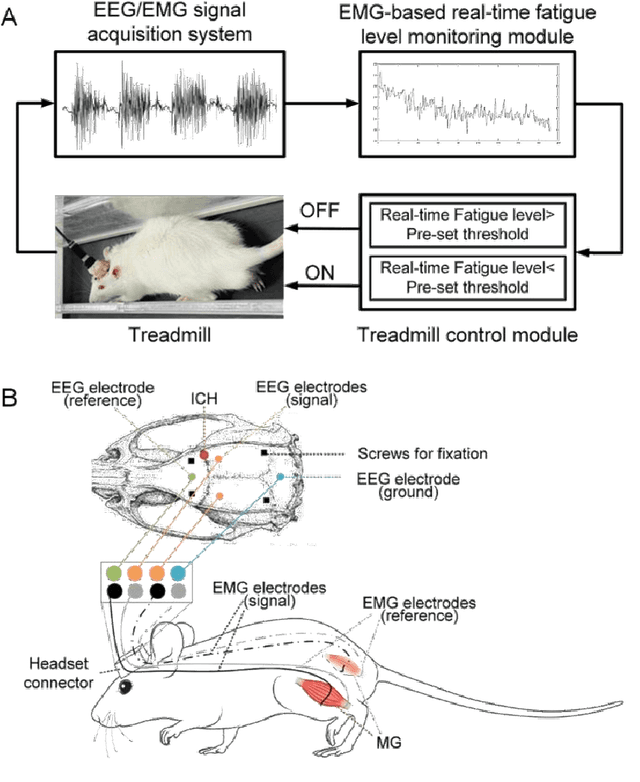
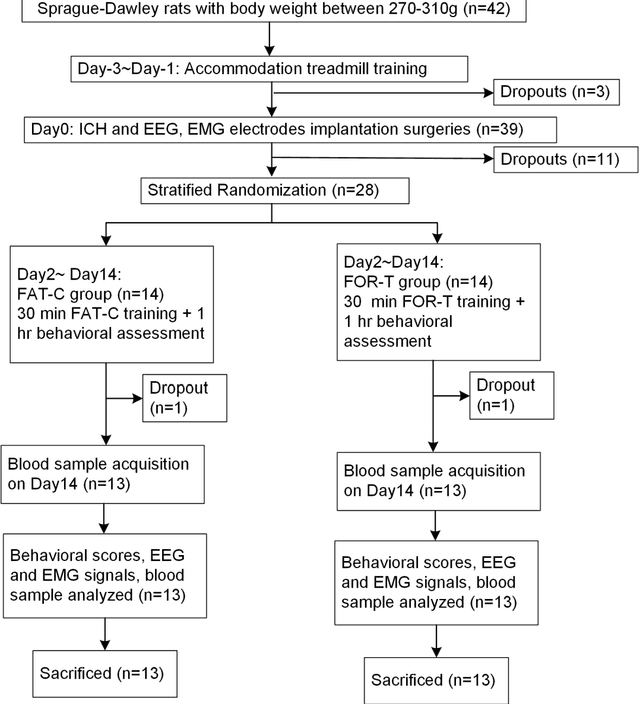
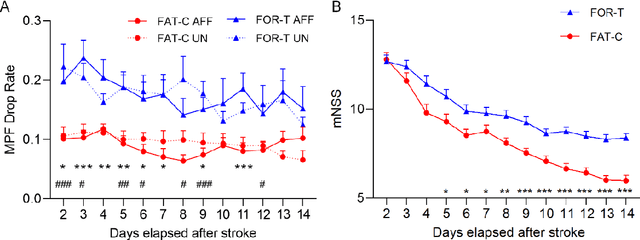
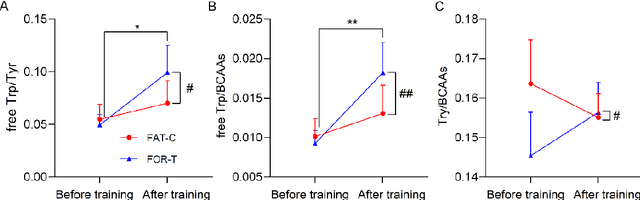
Abstract:Individualized training improved post-stroke motor function rehabilitation efficiency. However, the mechanisms of how individualized training facilitates recovery is not clear. This study explored the cortical and corticomuscular rehabilitative effects in post-stroke motor function recovery during individualized training. Sprague-Dawley rats with intracerebral hemorrhage (ICH) were randomly distributed into two groups: forced training (FOR-T, n=13) and individualized fatigue-controlled training (FAT-C, n=13) to receive training respectively from day 2 to day 14 post-stroke. The FAT-C group exhibited superior motor function recovery and less central fatigue compared to the FOR-T group. EEG PSD slope analysis demonstrated a better inter-hemispheric balance in FAT-C group compare to the FOR-T group. The dCMC analysis indicated that training-induced fatigue led to a short-term down-regulation of descending corticomuscular coherence (dCMC) and an up-regulation of ascending dCMC. In the long term, excessive fatigue hindered the recovery of descending control in the affected hemisphere. The individualized strategy of peripheral fatigue-controlled training achieved better motor function recovery, which could be attributed to the mitigation of central fatigue, optimization of inter-hemispheric balance and enhancement of descending control in the affected hemisphere.
Movable Antenna Array Aided Ultra Reliable Covert Communications
Dec 29, 2024



Abstract:In this paper, we construct a framework of the movable antenna (MA) aided covert communication shielded by the general noise uncertainty for the first time. According to the analysis performance on the derived closed-form expressions of the sum of the probabilities of the detection errors and the communication outage probability, the perfect covertness and the ultra reliability can be achieved by adjusting the antenna position in the MA array. Then, we formulate the communication covertness maximization problem with the constraints of the ultra reliability and the independent discrete movable position to optimize the transmitter's parameter. With the maximal ratio transmitting (MRT) design for the beamforming, we solve the closed-form optimal information transmit power and design a lightweight discrete projected gradient descent (DPGD) algorithm to determine the optimal antenna position. The numerical results show that the optimal achievable covertness and the feasible region of the steering angle with the MA array is significant larger than the one with the fixed-position antenna (FPA) array.
TopoCellGen: Generating Histopathology Cell Topology with a Diffusion Model
Dec 08, 2024Abstract:Accurately modeling multi-class cell topology is crucial in digital pathology, as it provides critical insights into tissue structure and pathology. The synthetic generation of cell topology enables realistic simulations of complex tissue environments, enhances downstream tasks by augmenting training data, aligns more closely with pathologists' domain knowledge, and offers new opportunities for controlling and generalizing the tumor microenvironment. In this paper, we propose a novel approach that integrates topological constraints into a diffusion model to improve the generation of realistic, contextually accurate cell topologies. Our method refines the simulation of cell distributions and interactions, increasing the precision and interpretability of results in downstream tasks such as cell detection and classification. To assess the topological fidelity of generated layouts, we introduce a new metric, Topological Frechet Distance (TopoFD), which overcomes the limitations of traditional metrics like FID in evaluating topological structure. Experimental results demonstrate the effectiveness of our approach in generating multi-class cell layouts that capture intricate topological relationships.
Learn2Synth: Learning Optimal Data Synthesis Using Hypergradients
Nov 23, 2024



Abstract:Domain randomization through synthesis is a powerful strategy to train networks that are unbiased with respect to the domain of the input images. Randomization allows networks to see a virtually infinite range of intensities and artifacts during training, thereby minimizing overfitting to appearance and maximizing generalization to unseen data. While powerful, this approach relies on the accurate tuning of a large set of hyper-parameters governing the probabilistic distribution of the synthesized images. Instead of manually tuning these parameters, we introduce Learn2Synth, a novel procedure in which synthesis parameters are learned using a small set of real labeled data. Unlike methods that impose constraints to align synthetic data with real data (e.g., contrastive or adversarial techniques), which risk misaligning the image and its label map, we tune an augmentation engine such that a segmentation network trained on synthetic data has optimal accuracy when applied to real data. This approach allows the training procedure to benefit from real labeled examples, without ever using these real examples to train the segmentation network, which avoids biasing the network towards the properties of the training set. Specifically, we develop both parametric and nonparametric strategies to augment the synthetic images, enhancing the segmentation network's performance. Experimental results on both synthetic and real-world datasets demonstrate the effectiveness of this learning strategy. Code is available at: https://github.com/HuXiaoling/Learn2Synth.
RankByGene: Gene-Guided Histopathology Representation Learning Through Cross-Modal Ranking Consistency
Nov 22, 2024Abstract:Spatial transcriptomics (ST) provides essential spatial context by mapping gene expression within tissue, enabling detailed study of cellular heterogeneity and tissue organization. However, aligning ST data with histology images poses challenges due to inherent spatial distortions and modality-specific variations. Existing methods largely rely on direct alignment, which often fails to capture complex cross-modal relationships. To address these limitations, we propose a novel framework that aligns gene and image features using a ranking-based alignment loss, preserving relative similarity across modalities and enabling robust multi-scale alignment. To further enhance the alignment's stability, we employ self-supervised knowledge distillation with a teacher-student network architecture, effectively mitigating disruptions from high dimensionality, sparsity, and noise in gene expression data. Extensive experiments on gene expression prediction and survival analysis demonstrate our framework's effectiveness, showing improved alignment and predictive performance over existing methods and establishing a robust tool for gene-guided image representation learning in digital pathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge