Aniruddha Ganguly
MERGE: Multi-faceted Hierarchical Graph-based GNN for Gene Expression Prediction from Whole Slide Histopathology Images
Dec 03, 2024



Abstract:Recent advances in Spatial Transcriptomics (ST) pair histology images with spatially resolved gene expression profiles, enabling predictions of gene expression across different tissue locations based on image patches. This opens up new possibilities for enhancing whole slide image (WSI) prediction tasks with localized gene expression. However, existing methods fail to fully leverage the interactions between different tissue locations, which are crucial for accurate joint prediction. To address this, we introduce MERGE (Multi-faceted hiErarchical gRaph for Gene Expressions), which combines a multi-faceted hierarchical graph construction strategy with graph neural networks (GNN) to improve gene expression predictions from WSIs. By clustering tissue image patches based on both spatial and morphological features, and incorporating intra- and inter-cluster edges, our approach fosters interactions between distant tissue locations during GNN learning. As an additional contribution, we evaluate different data smoothing techniques that are necessary to mitigate artifacts in ST data, often caused by technical imperfections. We advocate for adopting gene-aware smoothing methods that are more biologically justified. Experimental results on gene expression prediction show that our GNN method outperforms state-of-the-art techniques across multiple metrics.
RankByGene: Gene-Guided Histopathology Representation Learning Through Cross-Modal Ranking Consistency
Nov 22, 2024Abstract:Spatial transcriptomics (ST) provides essential spatial context by mapping gene expression within tissue, enabling detailed study of cellular heterogeneity and tissue organization. However, aligning ST data with histology images poses challenges due to inherent spatial distortions and modality-specific variations. Existing methods largely rely on direct alignment, which often fails to capture complex cross-modal relationships. To address these limitations, we propose a novel framework that aligns gene and image features using a ranking-based alignment loss, preserving relative similarity across modalities and enabling robust multi-scale alignment. To further enhance the alignment's stability, we employ self-supervised knowledge distillation with a teacher-student network architecture, effectively mitigating disruptions from high dimensionality, sparsity, and noise in gene expression data. Extensive experiments on gene expression prediction and survival analysis demonstrate our framework's effectiveness, showing improved alignment and predictive performance over existing methods and establishing a robust tool for gene-guided image representation learning in digital pathology.
3D-FFS: Faster 3D object detection with Focused Frustum Search in sensor fusion based networks
Mar 15, 2021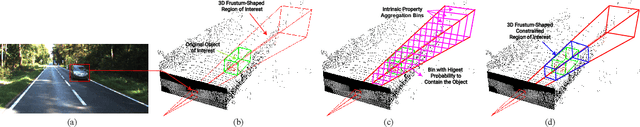
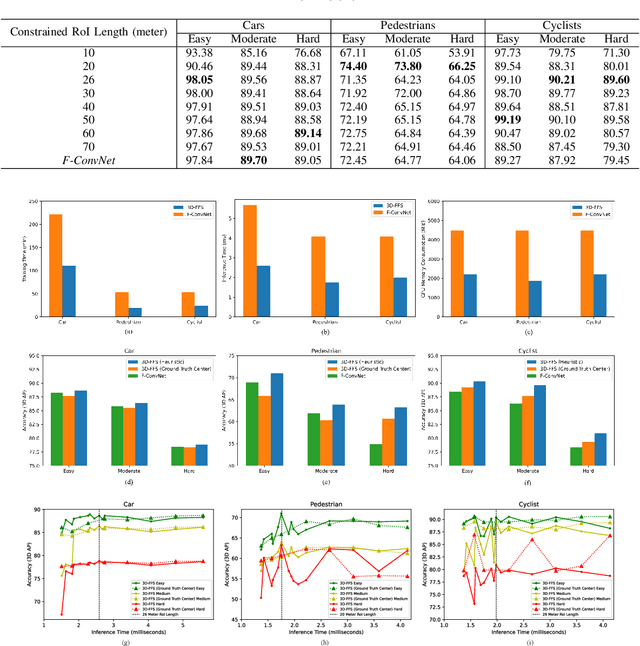
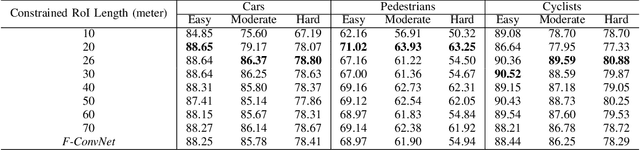
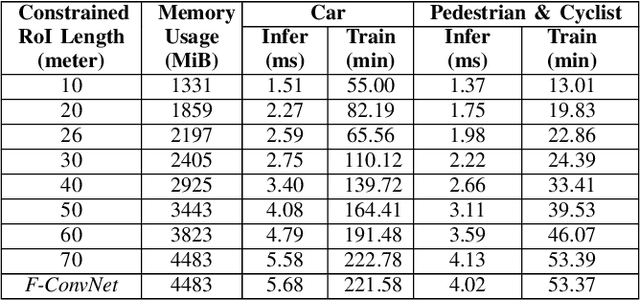
Abstract:In this work we propose 3D-FFS, a novel approach to make sensor fusion based 3D object detection networks significantly faster using a class of computationally inexpensive heuristics. Existing sensor fusion based networks generate 3D region proposals by leveraging inferences from 2D object detectors. However, as images have no depth information, these networks rely on extracting semantic features of points from the entire scene to locate the object. By leveraging aggregated intrinsic properties (e.g. point density) of the 3D point cloud data, 3D-FFS can substantially constrain the 3D search space and thereby significantly reduce training time, inference time and memory consumption without sacrificing accuracy. To demonstrate the efficacy of 3D-FFS, we have integrated it with Frustum ConvNet (F-ConvNet), a prominent sensor fusion based 3D object detection model. We assess the performance of 3D-FFS on the KITTI dataset. Compared to F-ConvNet, we achieve improvements in training and inference times by up to 62.84% and 56.46%, respectively, while reducing the memory usage by up to 58.53%. Additionally, we achieve 0.59%, 2.03% and 3.34% improvements in accuracy for the Car, Pedestrian and Cyclist classes, respectively. 3D-FFS shows a lot of promise in domains with limited computing power, such as autonomous vehicles, drones and robotics where LiDAR-Camera based sensor fusion perception systems are widely used.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge