Meilong Xu
MATCH: Multi-faceted Adaptive Topo-Consistency for Semi-Supervised Histopathology Segmentation
Oct 02, 2025



Abstract:In semi-supervised segmentation, capturing meaningful semantic structures from unlabeled data is essential. This is particularly challenging in histopathology image analysis, where objects are densely distributed. To address this issue, we propose a semi-supervised segmentation framework designed to robustly identify and preserve relevant topological features. Our method leverages multiple perturbed predictions obtained through stochastic dropouts and temporal training snapshots, enforcing topological consistency across these varied outputs. This consistency mechanism helps distinguish biologically meaningful structures from transient and noisy artifacts. A key challenge in this process is to accurately match the corresponding topological features across the predictions in the absence of ground truth. To overcome this, we introduce a novel matching strategy that integrates spatial overlap with global structural alignment, minimizing discrepancies among predictions. Extensive experiments demonstrate that our approach effectively reduces topological errors, resulting in more robust and accurate segmentations essential for reliable downstream analysis. Code is available at \href{https://github.com/Melon-Xu/MATCH}{https://github.com/Melon-Xu/MATCH}.
Towards Better Optimization For Listwise Preference in Diffusion Models
Oct 02, 2025



Abstract:Reinforcement learning from human feedback (RLHF) has proven effectiveness for aligning text-to-image (T2I) diffusion models with human preferences. Although Direct Preference Optimization (DPO) is widely adopted for its computational efficiency and avoidance of explicit reward modeling, its applications to diffusion models have primarily relied on pairwise preferences. The precise optimization of listwise preferences remains largely unaddressed. In practice, human feedback on image preferences often contains implicit ranked information, which conveys more precise human preferences than pairwise comparisons. In this work, we propose Diffusion-LPO, a simple and effective framework for Listwise Preference Optimization in diffusion models with listwise data. Given a caption, we aggregate user feedback into a ranked list of images and derive a listwise extension of the DPO objective under the Plackett-Luce model. Diffusion-LPO enforces consistency across the entire ranking by encouraging each sample to be preferred over all of its lower-ranked alternatives. We empirically demonstrate the effectiveness of Diffusion-LPO across various tasks, including text-to-image generation, image editing, and personalized preference alignment. Diffusion-LPO consistently outperforms pairwise DPO baselines on visual quality and preference alignment.
Bézier Meets Diffusion: Robust Generation Across Domains for Medical Image Segmentation
Sep 26, 2025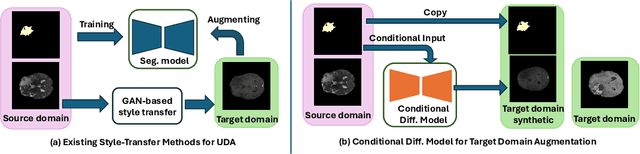
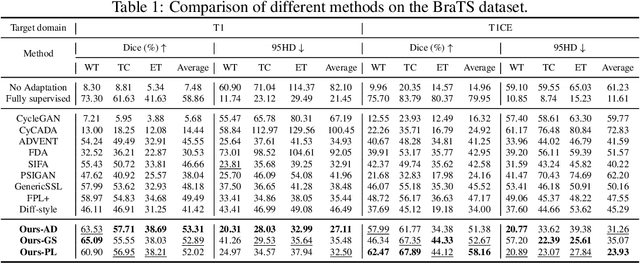
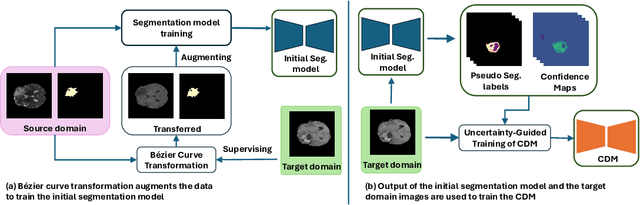
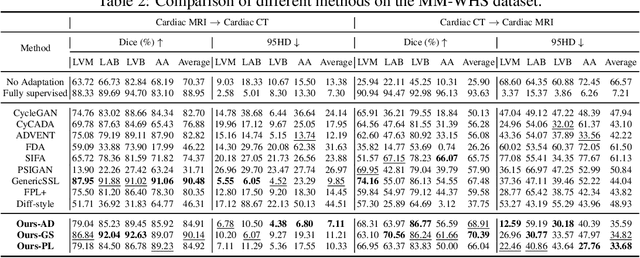
Abstract:Training robust learning algorithms across different medical imaging modalities is challenging due to the large domain gap. Unsupervised domain adaptation (UDA) mitigates this problem by using annotated images from the source domain and unlabeled images from the target domain to train the deep models. Existing approaches often rely on GAN-based style transfer, but these methods struggle to capture cross-domain mappings in regions with high variability. In this paper, we propose a unified framework, B\'ezier Meets Diffusion, for cross-domain image generation. First, we introduce a B\'ezier-curve-based style transfer strategy that effectively reduces the domain gap between source and target domains. The transferred source images enable the training of a more robust segmentation model across domains. Thereafter, using pseudo-labels generated by this segmentation model on the target domain, we train a conditional diffusion model (CDM) to synthesize high-quality, labeled target-domain images. To mitigate the impact of noisy pseudo-labels, we further develop an uncertainty-guided score matching method that improves the robustness of CDM training. Extensive experiments on public datasets demonstrate that our approach generates realistic labeled images, significantly augmenting the target domain and improving segmentation performance.
TopoCellGen: Generating Histopathology Cell Topology with a Diffusion Model
Dec 08, 2024Abstract:Accurately modeling multi-class cell topology is crucial in digital pathology, as it provides critical insights into tissue structure and pathology. The synthetic generation of cell topology enables realistic simulations of complex tissue environments, enhances downstream tasks by augmenting training data, aligns more closely with pathologists' domain knowledge, and offers new opportunities for controlling and generalizing the tumor microenvironment. In this paper, we propose a novel approach that integrates topological constraints into a diffusion model to improve the generation of realistic, contextually accurate cell topologies. Our method refines the simulation of cell distributions and interactions, increasing the precision and interpretability of results in downstream tasks such as cell detection and classification. To assess the topological fidelity of generated layouts, we introduce a new metric, Topological Frechet Distance (TopoFD), which overcomes the limitations of traditional metrics like FID in evaluating topological structure. Experimental results demonstrate the effectiveness of our approach in generating multi-class cell layouts that capture intricate topological relationships.
RankByGene: Gene-Guided Histopathology Representation Learning Through Cross-Modal Ranking Consistency
Nov 22, 2024Abstract:Spatial transcriptomics (ST) provides essential spatial context by mapping gene expression within tissue, enabling detailed study of cellular heterogeneity and tissue organization. However, aligning ST data with histology images poses challenges due to inherent spatial distortions and modality-specific variations. Existing methods largely rely on direct alignment, which often fails to capture complex cross-modal relationships. To address these limitations, we propose a novel framework that aligns gene and image features using a ranking-based alignment loss, preserving relative similarity across modalities and enabling robust multi-scale alignment. To further enhance the alignment's stability, we employ self-supervised knowledge distillation with a teacher-student network architecture, effectively mitigating disruptions from high dimensionality, sparsity, and noise in gene expression data. Extensive experiments on gene expression prediction and survival analysis demonstrate our framework's effectiveness, showing improved alignment and predictive performance over existing methods and establishing a robust tool for gene-guided image representation learning in digital pathology.
Spatial Diffusion for Cell Layout Generation
Sep 04, 2024



Abstract:Generative models, such as GANs and diffusion models, have been used to augment training sets and boost performances in different tasks. We focus on generative models for cell detection instead, i.e., locating and classifying cells in given pathology images. One important information that has been largely overlooked is the spatial patterns of the cells. In this paper, we propose a spatial-pattern-guided generative model for cell layout generation. Specifically, a novel diffusion model guided by spatial features and generates realistic cell layouts has been proposed. We explore different density models as spatial features for the diffusion model. In downstream tasks, we show that the generated cell layouts can be used to guide the generation of high-quality pathology images. Augmenting with these images can significantly boost the performance of SOTA cell detection methods. The code is available at https://github.com/superlc1995/Diffusion-cell.
TopoSemiSeg: Enforcing Topological Consistency for Semi-Supervised Segmentation of Histopathology Images
Dec 06, 2023Abstract:In computational pathology, segmenting densely distributed objects like glands and nuclei is crucial for downstream analysis. To alleviate the burden of obtaining pixel-wise annotations, semi-supervised learning methods learn from large amounts of unlabeled data. Nevertheless, existing semi-supervised methods overlook the topological information hidden in the unlabeled images and are thus prone to topological errors, e.g., missing or incorrectly merged/separated glands or nuclei. To address this issue, we propose TopoSemiSeg, the first semi-supervised method that learns the topological representation from unlabeled data. In particular, we propose a topology-aware teacher-student approach in which the teacher and student networks learn shared topological representations. To achieve this, we introduce topological consistency loss, which contains signal consistency and noise removal losses to ensure the learned representation is robust and focuses on true topological signals. Extensive experiments on public pathology image datasets show the superiority of our method, especially on topology-wise evaluation metrics. Code is available at https://github.com/Melon-Xu/TopoSemiSeg.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge