Wentao Huang
Debiasing Diffusion Priors via 3D Attention for Consistent Gaussian Splatting
Dec 16, 2025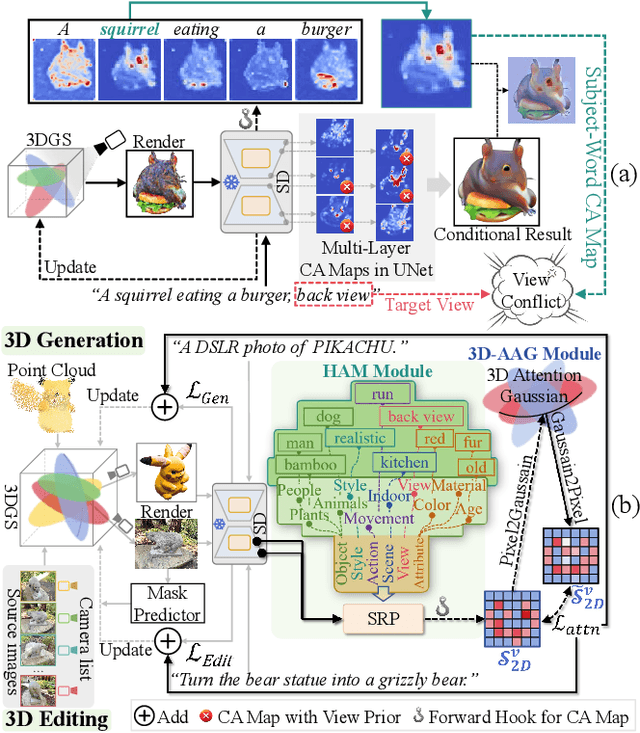
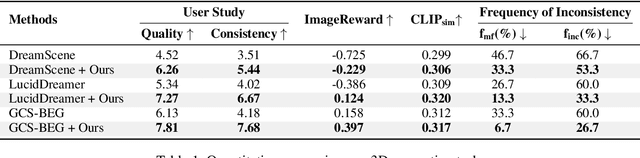
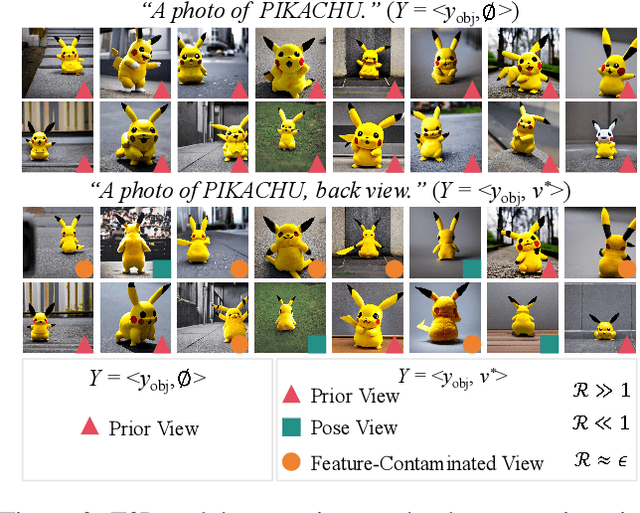
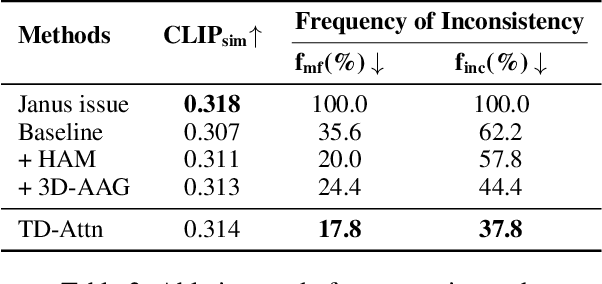
Abstract:Versatile 3D tasks (e.g., generation or editing) that distill from Text-to-Image (T2I) diffusion models have attracted significant research interest for not relying on extensive 3D training data. However, T2I models exhibit limitations resulting from prior view bias, which produces conflicting appearances between different views of an object. This bias causes subject-words to preferentially activate prior view features during cross-attention (CA) computation, regardless of the target view condition. To overcome this limitation, we conduct a comprehensive mathematical analysis to reveal the root cause of the prior view bias in T2I models. Moreover, we find different UNet layers show different effects of prior view in CA. Therefore, we propose a novel framework, TD-Attn, which addresses multi-view inconsistency via two key components: (1) the 3D-Aware Attention Guidance Module (3D-AAG) constructs a view-consistent 3D attention Gaussian for subject-words to enforce spatial consistency across attention-focused regions, thereby compensating for the limited spatial information in 2D individual view CA maps; (2) the Hierarchical Attention Modulation Module (HAM) utilizes a Semantic Guidance Tree (SGT) to direct the Semantic Response Profiler (SRP) in localizing and modulating CA layers that are highly responsive to view conditions, where the enhanced CA maps further support the construction of more consistent 3D attention Gaussians. Notably, HAM facilitates semantic-specific interventions, enabling controllable and precise 3D editing. Extensive experiments firmly establish that TD-Attn has the potential to serve as a universal plugin, significantly enhancing multi-view consistency across 3D tasks.
ReaSon: Reinforced Causal Search with Information Bottleneck for Video Understanding
Nov 16, 2025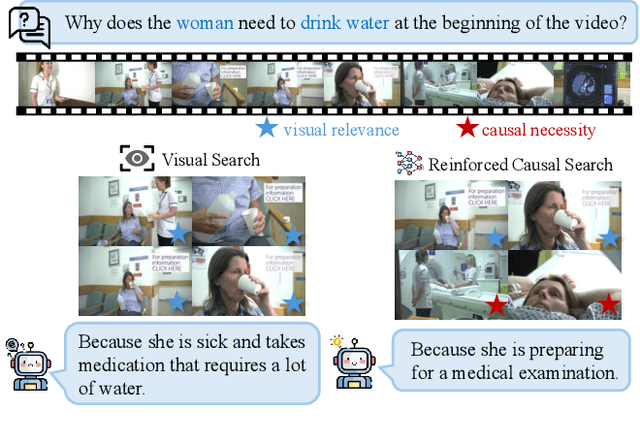
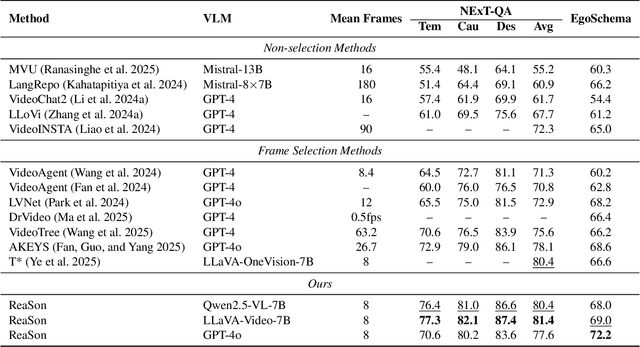
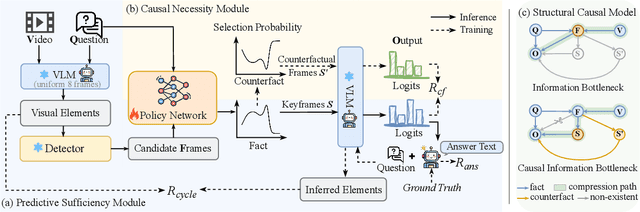
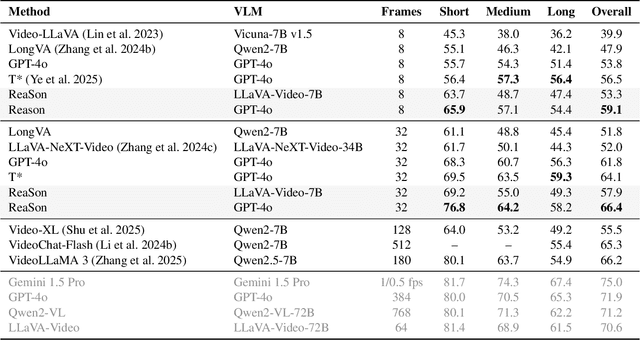
Abstract:Keyframe selection has become essential for video understanding with vision-language models (VLMs) due to limited input tokens and the temporal sparsity of relevant information across video frames. Video understanding often relies on effective keyframes that are not only informative but also causally decisive. To this end, we propose Reinforced Causal Search with Information Bottleneck (ReaSon), a framework that formulates keyframe selection as an optimization problem with the help of a novel Causal Information Bottleneck (CIB), which explicitly defines keyframes as those satisfying both predictive sufficiency and causal necessity. Specifically, ReaSon employs a learnable policy network to select keyframes from a visually relevant pool of candidate frames to capture predictive sufficiency, and then assesses causal necessity via counterfactual interventions. Finally, a composite reward aligned with the CIB principle is designed to guide the selection policy through reinforcement learning. Extensive experiments on NExT-QA, EgoSchema, and Video-MME demonstrate that ReaSon consistently outperforms existing state-of-the-art methods under limited-frame settings, validating its effectiveness and generalization ability.
Predict-then-Optimize for Seaport Power-Logistics Scheduling: Generalization across Varying Tasks Stream
Nov 13, 2025



Abstract:Power-logistics scheduling in modern seaports typically follow a predict-then-optimize pipeline. To enhance the decision quality of forecasts, decision-focused learning has been proposed, which aligns the training of forecasting models with downstream decision outcomes. However, this end-to-end design inherently restricts the value of forecasting models to only a specific task structure, and thus generalize poorly to evolving tasks induced by varying seaport vessel arrivals. We address this gap with a decision-focused continual learning framework that adapts online to a stream of scheduling tasks. Specifically, we introduce Fisher information based regularization to enhance cross-task generalization by preserving parameters critical to prior tasks. A differentiable convex surrogate is also developed to stabilize gradient backpropagation. The proposed approach enables learning a decision-aligned forecasting model across a varying tasks stream with a sustainable long-term computational burden. Experiments calibrated to the Jurong Port demonstrate superior decision performance and generalization over existing methods with reduced computational cost.
MERGE: Multi-faceted Hierarchical Graph-based GNN for Gene Expression Prediction from Whole Slide Histopathology Images
Dec 03, 2024



Abstract:Recent advances in Spatial Transcriptomics (ST) pair histology images with spatially resolved gene expression profiles, enabling predictions of gene expression across different tissue locations based on image patches. This opens up new possibilities for enhancing whole slide image (WSI) prediction tasks with localized gene expression. However, existing methods fail to fully leverage the interactions between different tissue locations, which are crucial for accurate joint prediction. To address this, we introduce MERGE (Multi-faceted hiErarchical gRaph for Gene Expressions), which combines a multi-faceted hierarchical graph construction strategy with graph neural networks (GNN) to improve gene expression predictions from WSIs. By clustering tissue image patches based on both spatial and morphological features, and incorporating intra- and inter-cluster edges, our approach fosters interactions between distant tissue locations during GNN learning. As an additional contribution, we evaluate different data smoothing techniques that are necessary to mitigate artifacts in ST data, often caused by technical imperfections. We advocate for adopting gene-aware smoothing methods that are more biologically justified. Experimental results on gene expression prediction show that our GNN method outperforms state-of-the-art techniques across multiple metrics.
RankByGene: Gene-Guided Histopathology Representation Learning Through Cross-Modal Ranking Consistency
Nov 22, 2024Abstract:Spatial transcriptomics (ST) provides essential spatial context by mapping gene expression within tissue, enabling detailed study of cellular heterogeneity and tissue organization. However, aligning ST data with histology images poses challenges due to inherent spatial distortions and modality-specific variations. Existing methods largely rely on direct alignment, which often fails to capture complex cross-modal relationships. To address these limitations, we propose a novel framework that aligns gene and image features using a ranking-based alignment loss, preserving relative similarity across modalities and enabling robust multi-scale alignment. To further enhance the alignment's stability, we employ self-supervised knowledge distillation with a teacher-student network architecture, effectively mitigating disruptions from high dimensionality, sparsity, and noise in gene expression data. Extensive experiments on gene expression prediction and survival analysis demonstrate our framework's effectiveness, showing improved alignment and predictive performance over existing methods and establishing a robust tool for gene-guided image representation learning in digital pathology.
A Simple Yet Effective Corpus Construction Framework for Indonesian Grammatical Error Correction
Oct 28, 2024Abstract:Currently, the majority of research in grammatical error correction (GEC) is concentrated on universal languages, such as English and Chinese. Many low-resource languages lack accessible evaluation corpora. How to efficiently construct high-quality evaluation corpora for GEC in low-resource languages has become a significant challenge. To fill these gaps, in this paper, we present a framework for constructing GEC corpora. Specifically, we focus on Indonesian as our research language and construct an evaluation corpus for Indonesian GEC using the proposed framework, addressing the limitations of existing evaluation corpora in Indonesian. Furthermore, we investigate the feasibility of utilizing existing large language models (LLMs), such as GPT-3.5-Turbo and GPT-4, to streamline corpus annotation efforts in GEC tasks. The results demonstrate significant potential for enhancing the performance of LLMs in low-resource language settings. Our code and corpus can be obtained from https://github.com/GKLMIP/GEC-Construction-Framework.
Hard Negative Sample Mining for Whole Slide Image Classification
Oct 03, 2024



Abstract:Weakly supervised whole slide image (WSI) classification is challenging due to the lack of patch-level labels and high computational costs. State-of-the-art methods use self-supervised patch-wise feature representations for multiple instance learning (MIL). Recently, methods have been proposed to fine-tune the feature representation on the downstream task using pseudo labeling, but mostly focusing on selecting high-quality positive patches. In this paper, we propose to mine hard negative samples during fine-tuning. This allows us to obtain better feature representations and reduce the training cost. Furthermore, we propose a novel patch-wise ranking loss in MIL to better exploit these hard negative samples. Experiments on two public datasets demonstrate the efficacy of these proposed ideas. Our codes are available at https://github.com/winston52/HNM-WSI
Convergence Analysis of Deep Residual Networks
May 13, 2022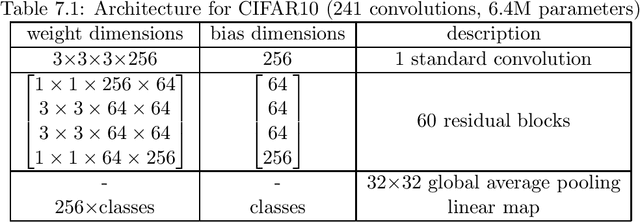
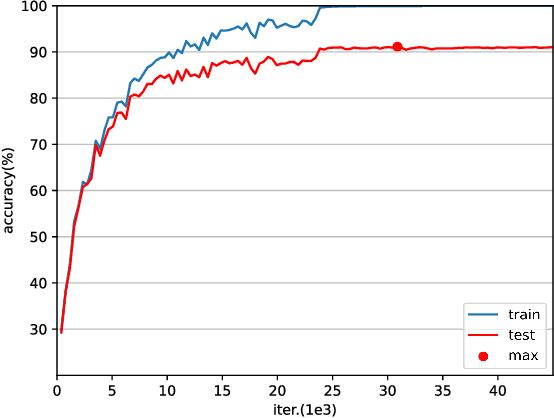
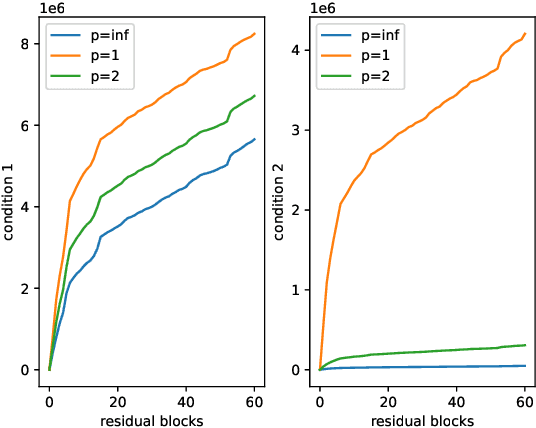
Abstract:Various powerful deep neural network architectures have made great contribution to the exciting successes of deep learning in the past two decades. Among them, deep Residual Networks (ResNets) are of particular importance because they demonstrated great usefulness in computer vision by winning the first place in many deep learning competitions. Also, ResNets were the first class of neural networks in the development history of deep learning that are really deep. It is of mathematical interest and practical meaning to understand the convergence of deep ResNets. We aim at characterizing the convergence of deep ResNets as the depth tends to infinity in terms of the parameters of the networks. Toward this purpose, we first give a matrix-vector description of general deep neural networks with shortcut connections and formulate an explicit expression for the networks by using the notions of activation domains and activation matrices. The convergence is then reduced to the convergence of two series involving infinite products of non-square matrices. By studying the two series, we establish a sufficient condition for pointwise convergence of ResNets. Our result is able to give justification for the design of ResNets. We also conduct experiments on benchmark machine learning data to verify our results.
Convergence of Deep Neural Networks with General Activation Functions and Pooling
May 13, 2022Abstract:Deep neural networks, as a powerful system to represent high dimensional complex functions, play a key role in deep learning. Convergence of deep neural networks is a fundamental issue in building the mathematical foundation for deep learning. We investigated the convergence of deep ReLU networks and deep convolutional neural networks in two recent researches (arXiv:2107.12530, 2109.13542). Only the Rectified Linear Unit (ReLU) activation was studied therein, and the important pooling strategy was not considered. In this current work, we study the convergence of deep neural networks as the depth tends to infinity for two other important activation functions: the leaky ReLU and the sigmoid function. Pooling will also be studied. As a result, we prove that the sufficient condition established in arXiv:2107.12530, 2109.13542 is still sufficient for the leaky ReLU networks. For contractive activation functions such as the sigmoid function, we establish a weaker sufficient condition for uniform convergence of deep neural networks.
Exploring Multi-dimensional Data via Subset Embedding
Apr 24, 2021



Abstract:Multi-dimensional data exploration is a classic research topic in visualization. Most existing approaches are designed for identifying record patterns in dimensional space or subspace. In this paper, we propose a visual analytics approach to exploring subset patterns. The core of the approach is a subset embedding network (SEN) that represents a group of subsets as uniformly-formatted embeddings. We implement the SEN as multiple subnets with separate loss functions. The design enables to handle arbitrary subsets and capture the similarity of subsets on single features, thus achieving accurate pattern exploration, which in most cases is searching for subsets having similar values on few features. Moreover, each subnet is a fully-connected neural network with one hidden layer. The simple structure brings high training efficiency. We integrate the SEN into a visualization system that achieves a 3-step workflow. Specifically, analysts (1) partition the given dataset into subsets, (2) select portions in a projected latent space created using the SEN, and (3) determine the existence of patterns within selected subsets. Generally, the system combines visualizations, interactions, automatic methods, and quantitative measures to balance the exploration flexibility and operation efficiency, and improve the interpretability and faithfulness of the identified patterns. Case studies and quantitative experiments on multiple open datasets demonstrate the general applicability and effectiveness of our approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge