Md. Shamsuzzoha Bayzid
3D-FFS: Faster 3D object detection with Focused Frustum Search in sensor fusion based networks
Mar 15, 2021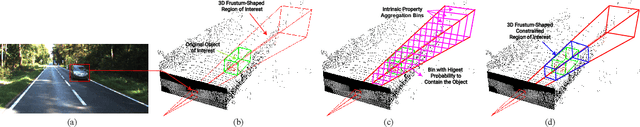
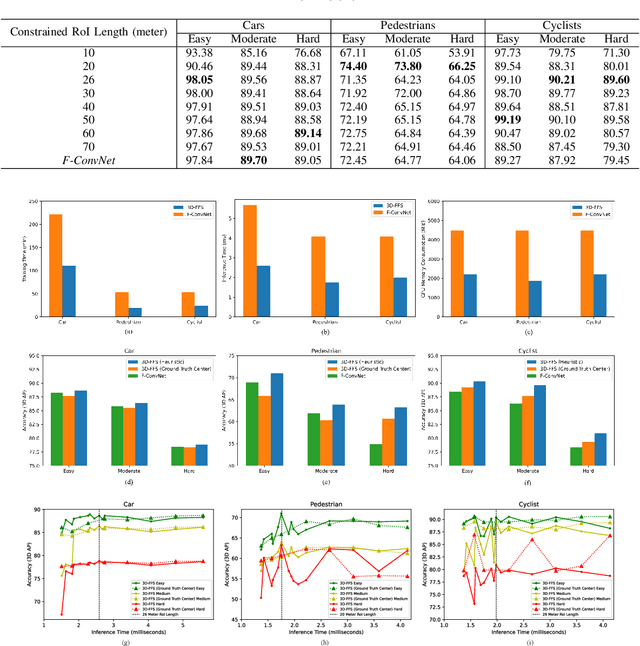
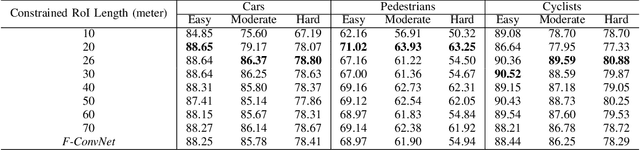
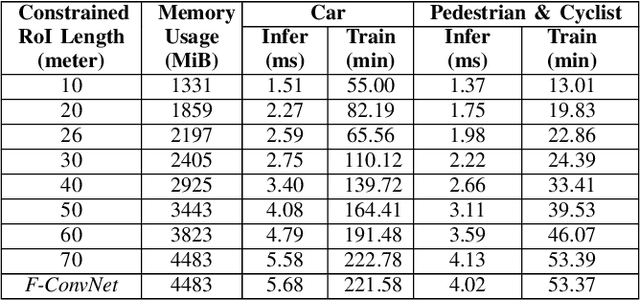
Abstract:In this work we propose 3D-FFS, a novel approach to make sensor fusion based 3D object detection networks significantly faster using a class of computationally inexpensive heuristics. Existing sensor fusion based networks generate 3D region proposals by leveraging inferences from 2D object detectors. However, as images have no depth information, these networks rely on extracting semantic features of points from the entire scene to locate the object. By leveraging aggregated intrinsic properties (e.g. point density) of the 3D point cloud data, 3D-FFS can substantially constrain the 3D search space and thereby significantly reduce training time, inference time and memory consumption without sacrificing accuracy. To demonstrate the efficacy of 3D-FFS, we have integrated it with Frustum ConvNet (F-ConvNet), a prominent sensor fusion based 3D object detection model. We assess the performance of 3D-FFS on the KITTI dataset. Compared to F-ConvNet, we achieve improvements in training and inference times by up to 62.84% and 56.46%, respectively, while reducing the memory usage by up to 58.53%. Additionally, we achieve 0.59%, 2.03% and 3.34% improvements in accuracy for the Car, Pedestrian and Cyclist classes, respectively. 3D-FFS shows a lot of promise in domains with limited computing power, such as autonomous vehicles, drones and robotics where LiDAR-Camera based sensor fusion perception systems are widely used.
Align-gram : Rethinking the Skip-gram Model for Protein Sequence Analysis
Dec 06, 2020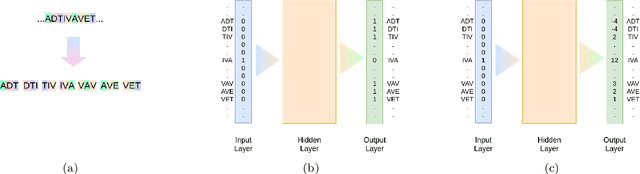
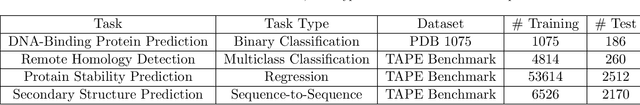
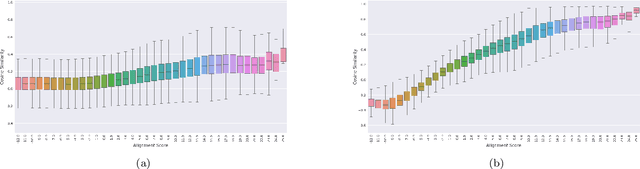
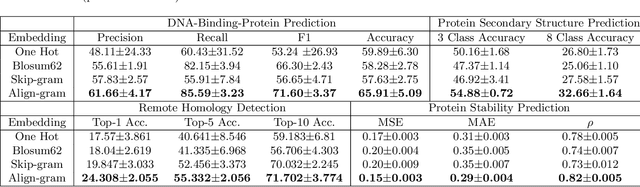
Abstract:Background: The inception of next generations sequencing technologies have exponentially increased the volume of biological sequence data. Protein sequences, being quoted as the `language of life', has been analyzed for a multitude of applications and inferences. Motivation: Owing to the rapid development of deep learning, in recent years there have been a number of breakthroughs in the domain of Natural Language Processing. Since these methods are capable of performing different tasks when trained with a sufficient amount of data, off-the-shelf models are used to perform various biological applications. In this study, we investigated the applicability of the popular Skip-gram model for protein sequence analysis and made an attempt to incorporate some biological insights into it. Results: We propose a novel $k$-mer embedding scheme, Align-gram, which is capable of mapping the similar $k$-mers close to each other in a vector space. Furthermore, we experiment with other sequence-based protein representations and observe that the embeddings derived from Align-gram aids modeling and training deep learning models better. Our experiments with a simple baseline LSTM model and a much complex CNN model of DeepGoPlus shows the potential of Align-gram in performing different types of deep learning applications for protein sequence analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge