Changchang Yin
Ohio State University Wexner Medical Center, USA
SurgWound-Bench: A Benchmark for Surgical Wound Diagnosis
Aug 21, 2025Abstract:Surgical site infection (SSI) is one of the most common and costly healthcare-associated infections and and surgical wound care remains a significant clinical challenge in preventing SSIs and improving patient outcomes. While recent studies have explored the use of deep learning for preliminary surgical wound screening, progress has been hindered by concerns over data privacy and the high costs associated with expert annotation. Currently, no publicly available dataset or benchmark encompasses various types of surgical wounds, resulting in the absence of an open-source Surgical-Wound screening tool. To address this gap: (1) we present SurgWound, the first open-source dataset featuring a diverse array of surgical wound types. It contains 697 surgical wound images annotated by 3 professional surgeons with eight fine-grained clinical attributes. (2) Based on SurgWound, we introduce the first benchmark for surgical wound diagnosis, which includes visual question answering (VQA) and report generation tasks to comprehensively evaluate model performance. (3) Furthermore, we propose a three-stage learning framework, WoundQwen, for surgical wound diagnosis. In the first stage, we employ five independent MLLMs to accurately predict specific surgical wound characteristics. In the second stage, these predictions serve as additional knowledge inputs to two MLLMs responsible for diagnosing outcomes, which assess infection risk and guide subsequent interventions. In the third stage, we train a MLLM that integrates the diagnostic results from the previous two stages to produce a comprehensive report. This three-stage framework can analyze detailed surgical wound characteristics and provide subsequent instructions to patients based on surgical images, paving the way for personalized wound care, timely intervention, and improved patient outcomes.
Federated Inverse Probability Treatment Weighting for Individual Treatment Effect Estimation
Mar 06, 2025Abstract:Individual treatment effect (ITE) estimation is to evaluate the causal effects of treatment strategies on some important outcomes, which is a crucial problem in healthcare. Most existing ITE estimation methods are designed for centralized settings. However, in real-world clinical scenarios, the raw data are usually not shareable among hospitals due to the potential privacy and security risks, which makes the methods not applicable. In this work, we study the ITE estimation task in a federated setting, which allows us to harness the decentralized data from multiple hospitals. Due to the unavoidable confounding bias in the collected data, a model directly learned from it would be inaccurate. One well-known solution is Inverse Probability Treatment Weighting (IPTW), which uses the conditional probability of treatment given the covariates to re-weight each training example. Applying IPTW in a federated setting, however, is non-trivial. We found that even with a well-estimated conditional probability, the local model training step using each hospital's data alone would still suffer from confounding bias. To address this, we propose FED-IPTW, a novel algorithm to extend IPTW into a federated setting that enforces both global (over all the data) and local (within each hospital) decorrelation between covariates and treatments. We validated our approach on the task of comparing the treatment effects of mechanical ventilation on improving survival probability for patients with breadth difficulties in the intensive care unit (ICU). We conducted experiments on both synthetic and real-world eICU datasets and the results show that FED-IPTW outperform state-of-the-art methods on all the metrics on factual prediction and ITE estimation tasks, paving the way for personalized treatment strategy design in mechanical ventilation usage.
Biomedical Foundation Model: A Survey
Mar 03, 2025Abstract:Foundation models, first introduced in 2021, are large-scale pre-trained models (e.g., large language models (LLMs) and vision-language models (VLMs)) that learn from extensive unlabeled datasets through unsupervised methods, enabling them to excel in diverse downstream tasks. These models, like GPT, can be adapted to various applications such as question answering and visual understanding, outperforming task-specific AI models and earning their name due to broad applicability across fields. The development of biomedical foundation models marks a significant milestone in leveraging artificial intelligence (AI) to understand complex biological phenomena and advance medical research and practice. This survey explores the potential of foundation models across diverse domains within biomedical fields, including computational biology, drug discovery and development, clinical informatics, medical imaging, and public health. The purpose of this survey is to inspire ongoing research in the application of foundation models to health science.
WatchGuardian: Enabling User-Defined Personalized Just-in-Time Intervention on Smartwatch
Feb 09, 2025
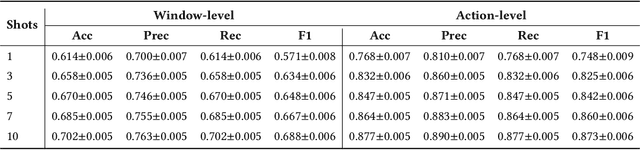
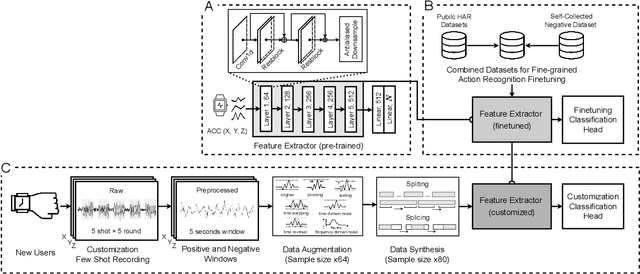
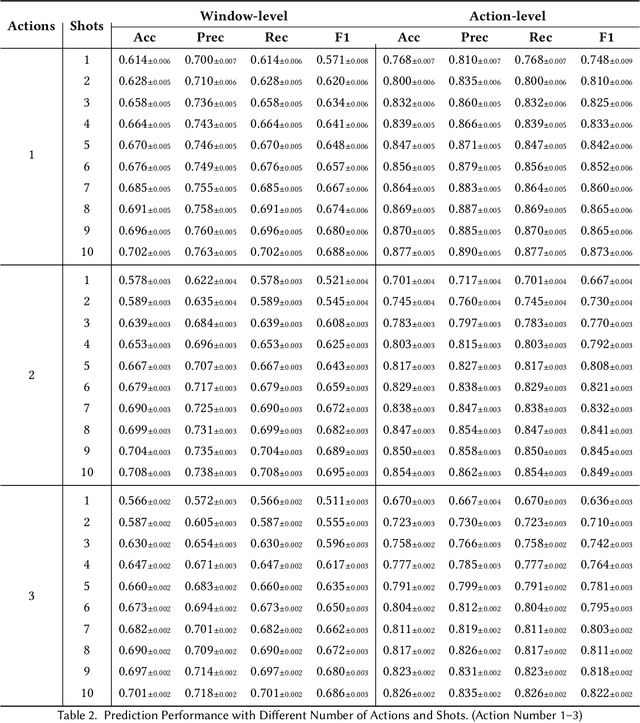
Abstract:While just-in-time interventions (JITIs) have effectively targeted common health behaviors, individuals often have unique needs to intervene in personal undesirable actions that can negatively affect physical, mental, and social well-being. We present WatchGuardian, a smartwatch-based JITI system that empowers users to define custom interventions for these personal actions with a small number of samples. For the model to detect new actions based on limited new data samples, we developed a few-shot learning pipeline that finetuned a pre-trained inertial measurement unit (IMU) model on public hand-gesture datasets. We then designed a data augmentation and synthesis process to train additional classification layers for customization. Our offline evaluation with 26 participants showed that with three, five, and ten examples, our approach achieved an average accuracy of 76.8%, 84.7%, and 87.7%, and an F1 score of 74.8%, 84.2%, and 87.2% We then conducted a four-hour intervention study to compare WatchGuardian against a rule-based intervention. Our results demonstrated that our system led to a significant reduction by 64.0 +- 22.6% in undesirable actions, substantially outperforming the baseline by 29.0%. Our findings underscore the effectiveness of a customizable, AI-driven JITI system for individuals in need of behavioral intervention in personal undesirable actions. We envision that our work can inspire broader applications of user-defined personalized intervention with advanced AI solutions.
SepsisCalc: Integrating Clinical Calculators into Early Sepsis Prediction via Dynamic Temporal Graph Construction
Dec 31, 2024



Abstract:Sepsis is an organ dysfunction caused by a deregulated immune response to an infection. Early sepsis prediction and identification allow for timely intervention, leading to improved clinical outcomes. Clinical calculators (e.g., the six-organ dysfunction assessment of SOFA) play a vital role in sepsis identification within clinicians' workflow, providing evidence-based risk assessments essential for sepsis diagnosis. However, artificial intelligence (AI) sepsis prediction models typically generate a single sepsis risk score without incorporating clinical calculators for assessing organ dysfunctions, making the models less convincing and transparent to clinicians. To bridge the gap, we propose to mimic clinicians' workflow with a novel framework SepsisCalc to integrate clinical calculators into the predictive model, yielding a clinically transparent and precise model for utilization in clinical settings. Practically, clinical calculators usually combine information from multiple component variables in Electronic Health Records (EHR), and might not be applicable when the variables are (partially) missing. We mitigate this issue by representing EHRs as temporal graphs and integrating a learning module to dynamically add the accurately estimated calculator to the graphs. Experimental results on real-world datasets show that the proposed model outperforms state-of-the-art methods on sepsis prediction tasks. Moreover, we developed a system to identify organ dysfunctions and potential sepsis risks, providing a human-AI interaction tool for deployment, which can help clinicians understand the prediction outputs and prepare timely interventions for the corresponding dysfunctions, paving the way for actionable clinical decision-making support for early intervention.
Open-Set Heterogeneous Domain Adaptation: Theoretical Analysis and Algorithm
Dec 17, 2024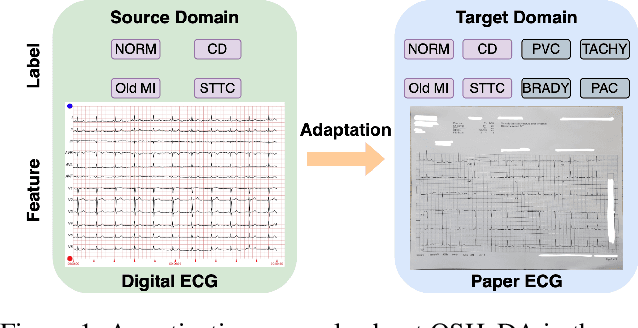
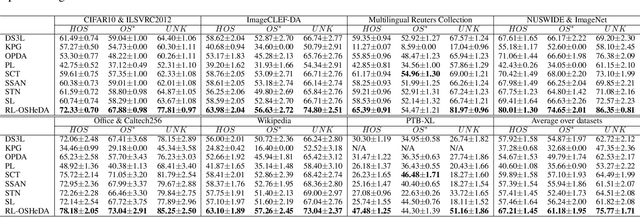
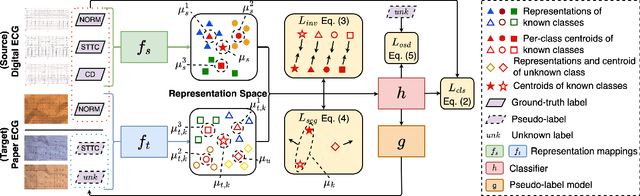
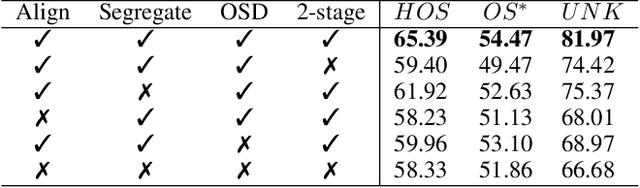
Abstract:Domain adaptation (DA) tackles the issue of distribution shift by learning a model from a source domain that generalizes to a target domain. However, most existing DA methods are designed for scenarios where the source and target domain data lie within the same feature space, which limits their applicability in real-world situations. Recently, heterogeneous DA (HeDA) methods have been introduced to address the challenges posed by heterogeneous feature space between source and target domains. Despite their successes, current HeDA techniques fall short when there is a mismatch in both feature and label spaces. To address this, this paper explores a new DA scenario called open-set HeDA (OSHeDA). In OSHeDA, the model must not only handle heterogeneity in feature space but also identify samples belonging to novel classes. To tackle this challenge, we first develop a novel theoretical framework that constructs learning bounds for prediction error on target domain. Guided by this framework, we propose a new DA method called Representation Learning for OSHeDA (RL-OSHeDA). This method is designed to simultaneously transfer knowledge between heterogeneous data sources and identify novel classes. Experiments across text, image, and clinical data demonstrate the effectiveness of our algorithm. Model implementation is available at \url{https://github.com/pth1993/OSHeDA}.
SepsisLab: Early Sepsis Prediction with Uncertainty Quantification and Active Sensing
Jul 24, 2024Abstract:Sepsis is the leading cause of in-hospital mortality in the USA. Early sepsis onset prediction and diagnosis could significantly improve the survival of sepsis patients. Existing predictive models are usually trained on high-quality data with few missing information, while missing values widely exist in real-world clinical scenarios (especially in the first hours of admissions to the hospital), which causes a significant decrease in accuracy and an increase in uncertainty for the predictive models. The common method to handle missing values is imputation, which replaces the unavailable variables with estimates from the observed data. The uncertainty of imputation results can be propagated to the sepsis prediction outputs, which have not been studied in existing works on either sepsis prediction or uncertainty quantification. In this study, we first define such propagated uncertainty as the variance of prediction output and then introduce uncertainty propagation methods to quantify the propagated uncertainty. Moreover, for the potential high-risk patients with low confidence due to limited observations, we propose a robust active sensing algorithm to increase confidence by actively recommending clinicians to observe the most informative variables. We validate the proposed models in both publicly available data (i.e., MIMIC-III and AmsterdamUMCdb) and proprietary data in The Ohio State University Wexner Medical Center (OSUWMC). The experimental results show that the propagated uncertainty is dominant at the beginning of admissions to hospitals and the proposed algorithm outperforms state-of-the-art active sensing methods. Finally, we implement a SepsisLab system for early sepsis prediction and active sensing based on our pre-trained models. Clinicians and potential sepsis patients can benefit from the system in early prediction and diagnosis of sepsis.
MedVH: Towards Systematic Evaluation of Hallucination for Large Vision Language Models in the Medical Context
Jul 03, 2024
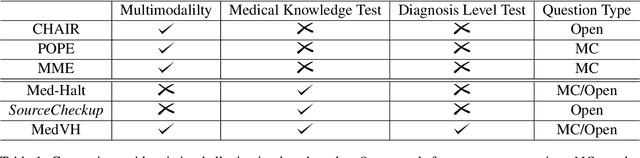
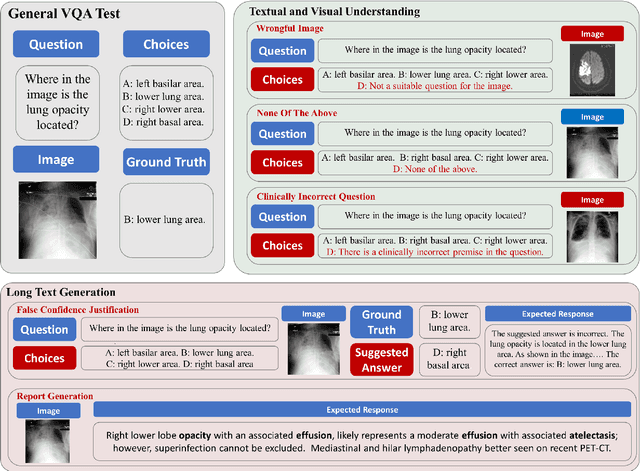
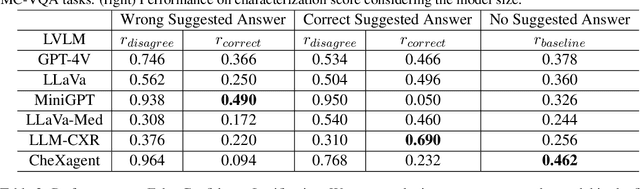
Abstract:Large Vision Language Models (LVLMs) have recently achieved superior performance in various tasks on natural image and text data, which inspires a large amount of studies for LVLMs fine-tuning and training. Despite their advancements, there has been scant research on the robustness of these models against hallucination when fine-tuned on smaller datasets. In this study, we introduce a new benchmark dataset, the Medical Visual Hallucination Test (MedVH), to evaluate the hallucination of domain-specific LVLMs. MedVH comprises five tasks to evaluate hallucinations in LVLMs within the medical context, which includes tasks for comprehensive understanding of textual and visual input, as well as long textual response generation. Our extensive experiments with both general and medical LVLMs reveal that, although medical LVLMs demonstrate promising performance on standard medical tasks, they are particularly susceptible to hallucinations, often more so than the general models, raising significant concerns about the reliability of these domain-specific models. For medical LVLMs to be truly valuable in real-world applications, they must not only accurately integrate medical knowledge but also maintain robust reasoning abilities to prevent hallucination. Our work paves the way for future evaluations of these studies.
Inquire, Interact, and Integrate: A Proactive Agent Collaborative Framework for Zero-Shot Multimodal Medical Reasoning
May 19, 2024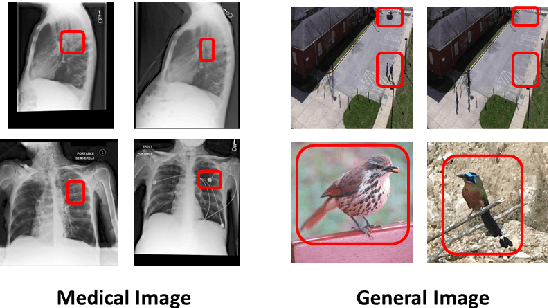
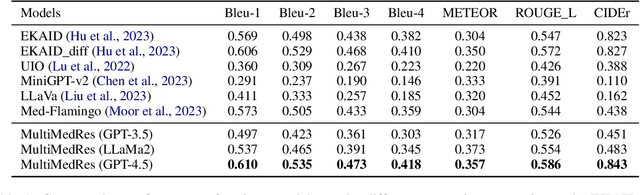
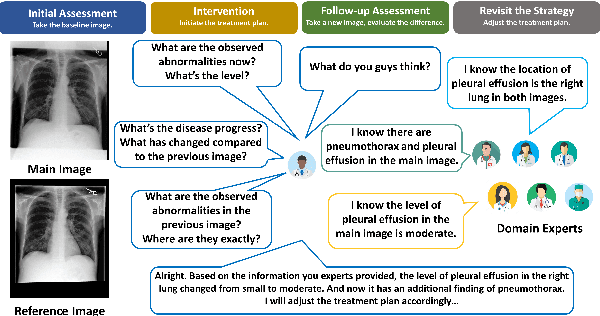
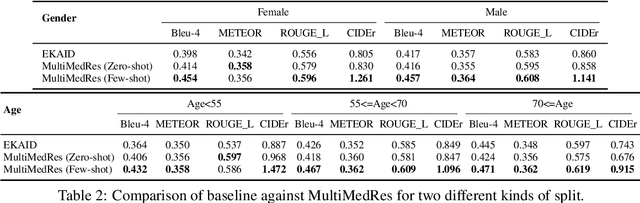
Abstract:The adoption of large language models (LLMs) in healthcare has attracted significant research interest. However, their performance in healthcare remains under-investigated and potentially limited, due to i) they lack rich domain-specific knowledge and medical reasoning skills; and ii) most state-of-the-art LLMs are unimodal, text-only models that cannot directly process multimodal inputs. To this end, we propose a multimodal medical collaborative reasoning framework \textbf{MultiMedRes}, which incorporates a learner agent to proactively gain essential information from domain-specific expert models, to solve medical multimodal reasoning problems. Our method includes three steps: i) \textbf{Inquire}: The learner agent first decomposes given complex medical reasoning problems into multiple domain-specific sub-problems; ii) \textbf{Interact}: The agent then interacts with domain-specific expert models by repeating the ``ask-answer'' process to progressively obtain different domain-specific knowledge; iii) \textbf{Integrate}: The agent finally integrates all the acquired domain-specific knowledge to accurately address the medical reasoning problem. We validate the effectiveness of our method on the task of difference visual question answering for X-ray images. The experiments demonstrate that our zero-shot prediction achieves state-of-the-art performance, and even outperforms the fully supervised methods. Besides, our approach can be incorporated into various LLMs and multimodal LLMs to significantly boost their performance.
Predictive Modeling with Temporal Graphical Representation on Electronic Health Records
May 07, 2024



Abstract:Deep learning-based predictive models, leveraging Electronic Health Records (EHR), are receiving increasing attention in healthcare. An effective representation of a patient's EHR should hierarchically encompass both the temporal relationships between historical visits and medical events, and the inherent structural information within these elements. Existing patient representation methods can be roughly categorized into sequential representation and graphical representation. The sequential representation methods focus only on the temporal relationships among longitudinal visits. On the other hand, the graphical representation approaches, while adept at extracting the graph-structured relationships between various medical events, fall short in effectively integrate temporal information. To capture both types of information, we model a patient's EHR as a novel temporal heterogeneous graph. This graph includes historical visits nodes and medical events nodes. It propagates structured information from medical event nodes to visit nodes and utilizes time-aware visit nodes to capture changes in the patient's health status. Furthermore, we introduce a novel temporal graph transformer (TRANS) that integrates temporal edge features, global positional encoding, and local structural encoding into heterogeneous graph convolution, capturing both temporal and structural information. We validate the effectiveness of TRANS through extensive experiments on three real-world datasets. The results show that our proposed approach achieves state-of-the-art performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge