Yuanlong Wang
Achieving Fairness Without Harm via Selective Demographic Experts
Nov 09, 2025Abstract:As machine learning systems become increasingly integrated into human-centered domains such as healthcare, ensuring fairness while maintaining high predictive performance is critical. Existing bias mitigation techniques often impose a trade-off between fairness and accuracy, inadvertently degrading performance for certain demographic groups. In high-stakes domains like clinical diagnosis, such trade-offs are ethically and practically unacceptable. In this study, we propose a fairness-without-harm approach by learning distinct representations for different demographic groups and selectively applying demographic experts consisting of group-specific representations and personalized classifiers through a no-harm constrained selection. We evaluate our approach on three real-world medical datasets -- covering eye disease, skin cancer, and X-ray diagnosis -- as well as two face datasets. Extensive empirical results demonstrate the effectiveness of our approach in achieving fairness without harm.
Memorization $ eq$ Understanding: Do Large Language Models Have the Ability of Scenario Cognition?
Sep 05, 2025Abstract:Driven by vast and diverse textual data, large language models (LLMs) have demonstrated impressive performance across numerous natural language processing (NLP) tasks. Yet, a critical question persists: does their generalization arise from mere memorization of training data or from deep semantic understanding? To investigate this, we propose a bi-perspective evaluation framework to assess LLMs' scenario cognition - the ability to link semantic scenario elements with their arguments in context. Specifically, we introduce a novel scenario-based dataset comprising diverse textual descriptions of fictional facts, annotated with scenario elements. LLMs are evaluated through their capacity to answer scenario-related questions (model output perspective) and via probing their internal representations for encoded scenario elements-argument associations (internal representation perspective). Our experiments reveal that current LLMs predominantly rely on superficial memorization, failing to achieve robust semantic scenario cognition, even in simple cases. These findings expose critical limitations in LLMs' semantic understanding and offer cognitive insights for advancing their capabilities.
Open-Set Heterogeneous Domain Adaptation: Theoretical Analysis and Algorithm
Dec 17, 2024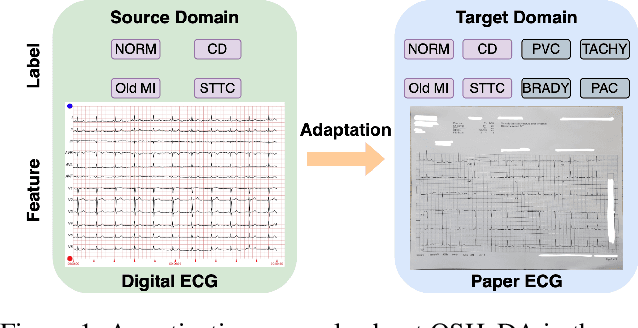
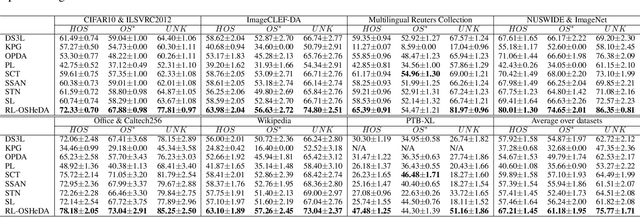
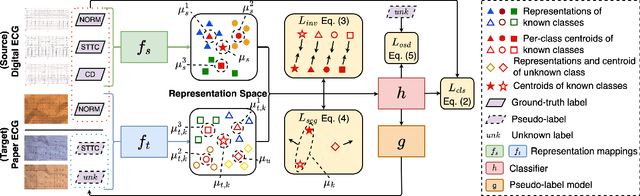
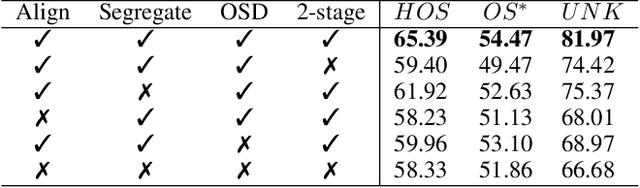
Abstract:Domain adaptation (DA) tackles the issue of distribution shift by learning a model from a source domain that generalizes to a target domain. However, most existing DA methods are designed for scenarios where the source and target domain data lie within the same feature space, which limits their applicability in real-world situations. Recently, heterogeneous DA (HeDA) methods have been introduced to address the challenges posed by heterogeneous feature space between source and target domains. Despite their successes, current HeDA techniques fall short when there is a mismatch in both feature and label spaces. To address this, this paper explores a new DA scenario called open-set HeDA (OSHeDA). In OSHeDA, the model must not only handle heterogeneity in feature space but also identify samples belonging to novel classes. To tackle this challenge, we first develop a novel theoretical framework that constructs learning bounds for prediction error on target domain. Guided by this framework, we propose a new DA method called Representation Learning for OSHeDA (RL-OSHeDA). This method is designed to simultaneously transfer knowledge between heterogeneous data sources and identify novel classes. Experiments across text, image, and clinical data demonstrate the effectiveness of our algorithm. Model implementation is available at \url{https://github.com/pth1993/OSHeDA}.
Predictive Modeling with Temporal Graphical Representation on Electronic Health Records
May 07, 2024



Abstract:Deep learning-based predictive models, leveraging Electronic Health Records (EHR), are receiving increasing attention in healthcare. An effective representation of a patient's EHR should hierarchically encompass both the temporal relationships between historical visits and medical events, and the inherent structural information within these elements. Existing patient representation methods can be roughly categorized into sequential representation and graphical representation. The sequential representation methods focus only on the temporal relationships among longitudinal visits. On the other hand, the graphical representation approaches, while adept at extracting the graph-structured relationships between various medical events, fall short in effectively integrate temporal information. To capture both types of information, we model a patient's EHR as a novel temporal heterogeneous graph. This graph includes historical visits nodes and medical events nodes. It propagates structured information from medical event nodes to visit nodes and utilizes time-aware visit nodes to capture changes in the patient's health status. Furthermore, we introduce a novel temporal graph transformer (TRANS) that integrates temporal edge features, global positional encoding, and local structural encoding into heterogeneous graph convolution, capturing both temporal and structural information. We validate the effectiveness of TRANS through extensive experiments on three real-world datasets. The results show that our proposed approach achieves state-of-the-art performance.
On compression rate of quantum autoencoders: Control design, numerical and experimental realization
May 22, 2020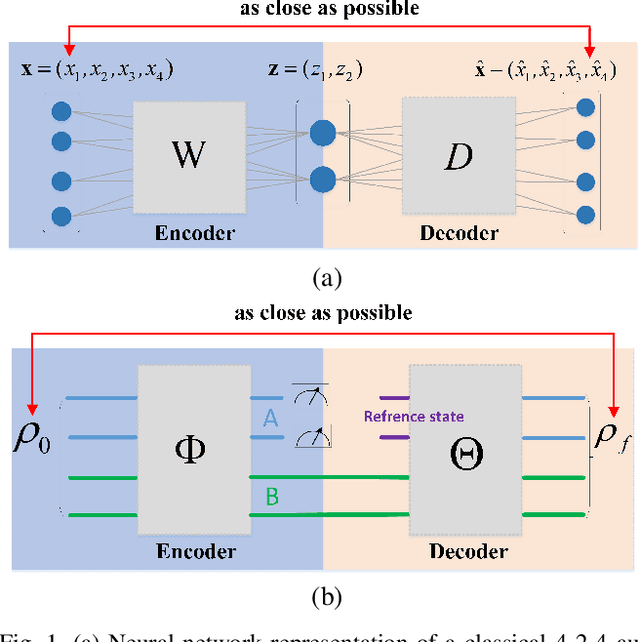
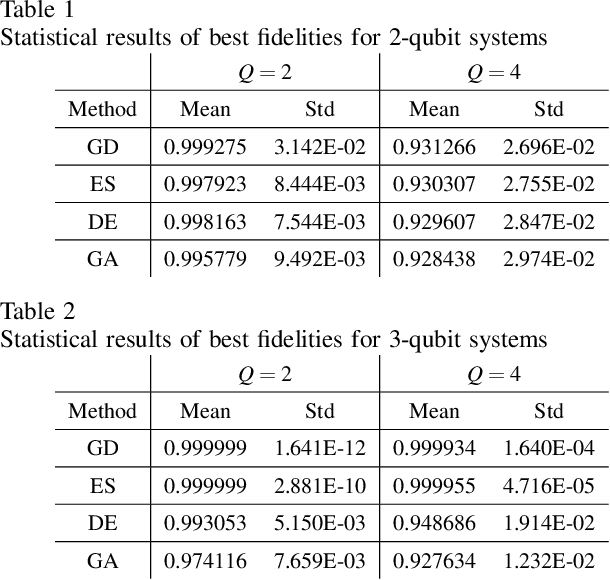
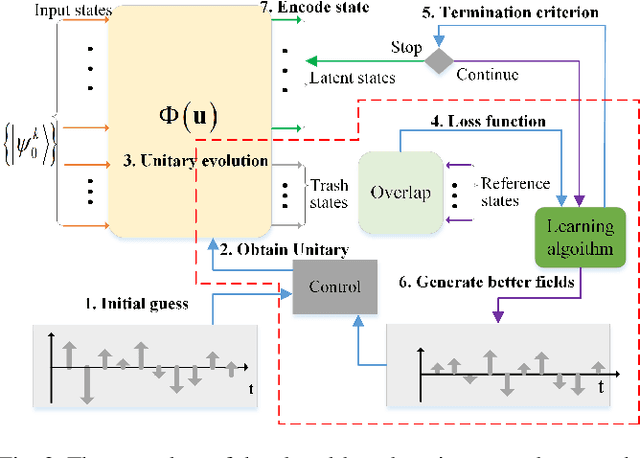
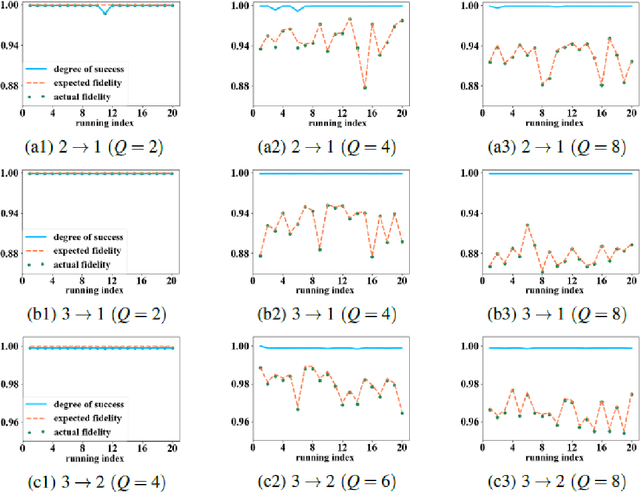
Abstract:Quantum autoencoders which aim at compressing quantum information in a low-dimensional latent space lie in the heart of automatic data compression in the field of quantum information. In this paper, we establish an upper bound of the compression rate for a given quantum autoencoder and present a learning control approach for training the autoencoder to achieve the maximal compression rate. The upper bound of the compression rate is theoretically proven using eigen-decomposition and matrix differentiation, which is determined by the eigenvalues of the density matrix representation of the input states. Numerical results on 2-qubit and 3-qubit systems are presented to demonstrate how to train the quantum autoencoder to achieve the theoretically maximal compression, and the training performance using different machine learning algorithms is compared. Experimental results of a quantum autoencoder using quantum optical systems are illustrated for compressing two 2-qubit states into two 1-qubit states.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge