Sheng Wang
Molecular Representations in Implicit Functional Space via Hyper-Networks
Jan 29, 2026Abstract:Molecular representations fundamentally shape how machine learning systems reason about molecular structure and physical properties. Most existing approaches adopt a discrete pipeline: molecules are encoded as sequences, graphs, or point clouds, mapped to fixed-dimensional embeddings, and then used for task-specific prediction. This paradigm treats molecules as discrete objects, despite their intrinsically continuous and field-like physical nature. We argue that molecular learning can instead be formulated as learning in function space. Specifically, we model each molecule as a continuous function over three-dimensional (3D) space and treat this molecular field as the primary object of representation. From this perspective, conventional molecular representations arise as particular sampling schemes of an underlying continuous object. We instantiate this formulation with MolField, a hyper-network-based framework that learns distributions over molecular fields. To ensure physical consistency, these functions are defined over canonicalized coordinates, yielding invariance to global SE(3) transformations. To enable learning directly over functions, we introduce a structured weight tokenization and train a sequence-based hyper-network to model a shared prior over molecular fields. We evaluate MolField on molecular dynamics and property prediction. Our results show that treating molecules as continuous functions fundamentally changes how molecular representations generalize across tasks and yields downstream behavior that is stable to how molecules are discretized or queried.
Adaptation of Agentic AI
Dec 22, 2025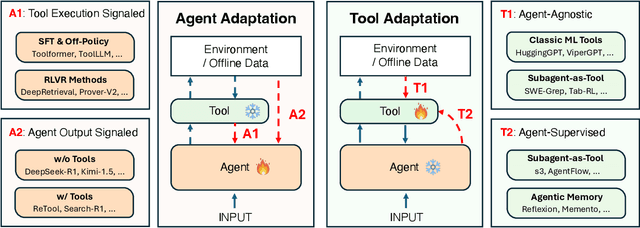
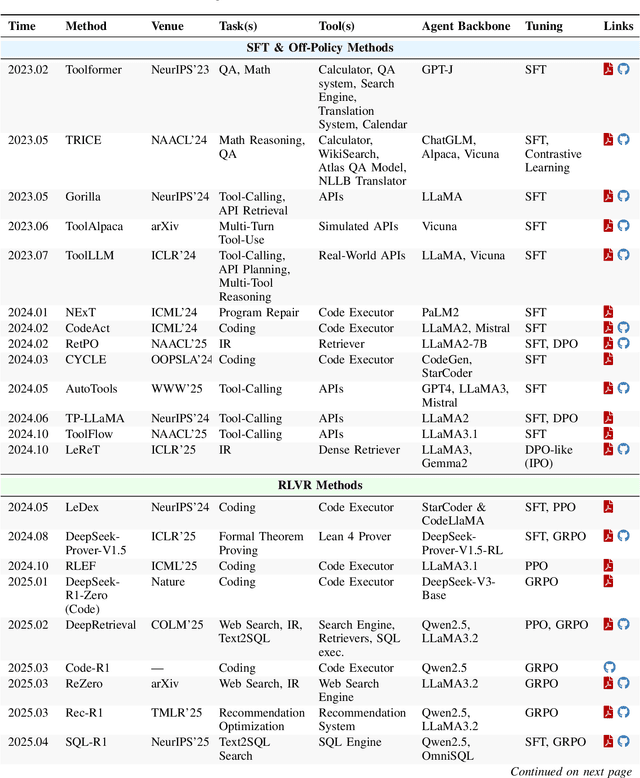
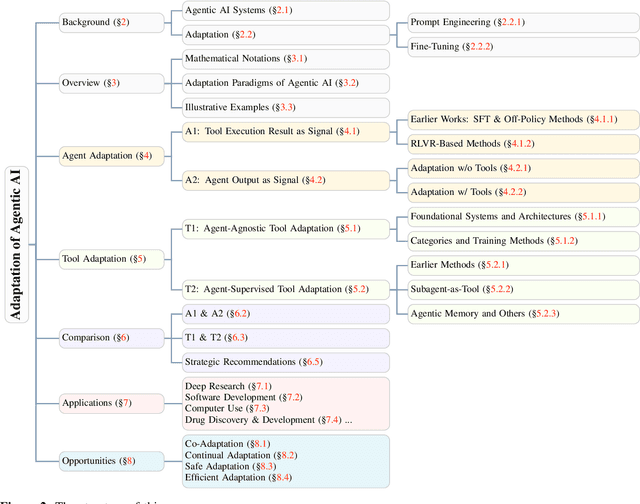
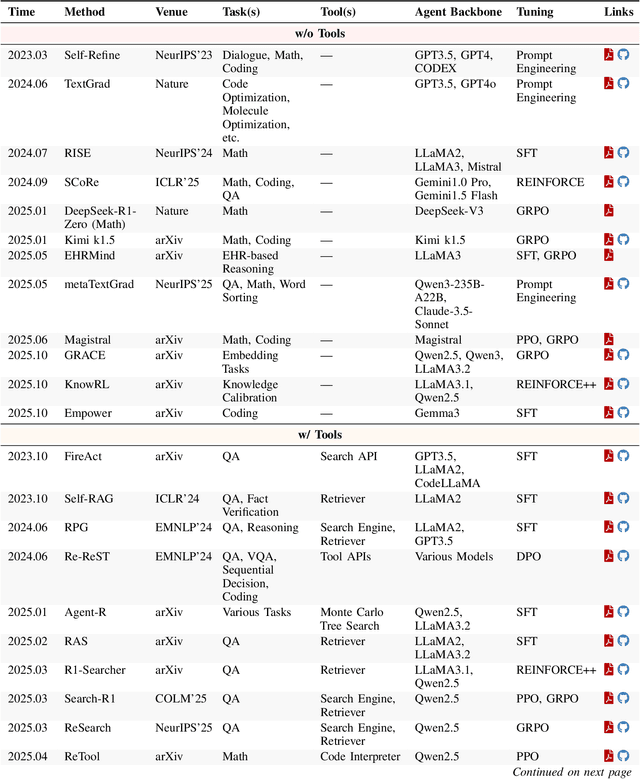
Abstract:Cutting-edge agentic AI systems are built on foundation models that can be adapted to plan, reason, and interact with external tools to perform increasingly complex and specialized tasks. As these systems grow in capability and scope, adaptation becomes a central mechanism for improving performance, reliability, and generalization. In this paper, we unify the rapidly expanding research landscape into a systematic framework that spans both agent adaptations and tool adaptations. We further decompose these into tool-execution-signaled and agent-output-signaled forms of agent adaptation, as well as agent-agnostic and agent-supervised forms of tool adaptation. We demonstrate that this framework helps clarify the design space of adaptation strategies in agentic AI, makes their trade-offs explicit, and provides practical guidance for selecting or switching among strategies during system design. We then review the representative approaches in each category, analyze their strengths and limitations, and highlight key open challenges and future opportunities. Overall, this paper aims to offer a conceptual foundation and practical roadmap for researchers and practitioners seeking to build more capable, efficient, and reliable agentic AI systems.
Task Vector in TTS: Toward Emotionally Expressive Dialectal Speech Synthesis
Dec 21, 2025Abstract:Recent advances in text-to-speech (TTS) have yielded remarkable improvements in naturalness and intelligibility. Building on these achievements, research has increasingly shifted toward enhancing the expressiveness of generated speech, such as dialectal and emotional TTS. However, cross-style synthesis combining both dialect and emotion remains challenging and largely unexplored, mainly due to the scarcity of dialectal data with emotional labels. To address this, we propose Hierarchical Expressive Vector (HE-Vector), a two-stage method for Emotional Dialectal TTS. In the first stage, we construct different task vectors to model dialectal and emotional styles independently, and then enhance single-style synthesis by adjusting their weights, a method we refer to as Expressive Vector (E-Vector). For the second stage, we hierarchically integrate these vectors to achieve controllable emotionally expressive dialect synthesis without requiring jointly labeled data, corresponding to Hierarchical Expressive Vector (HE-Vector). Experimental results demonstrate that HE-Vectors achieve superior performance in dialect synthesis, and promising results in synthesizing emotionally expressive dialectal speech in a zero-shot setting.
FutureX: Enhance End-to-End Autonomous Driving via Latent Chain-of-Thought World Model
Dec 12, 2025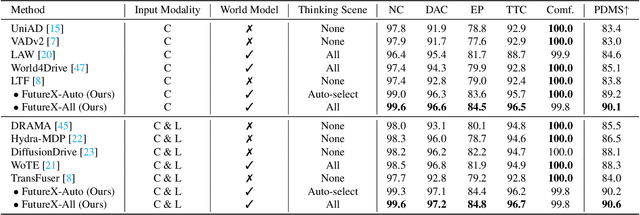
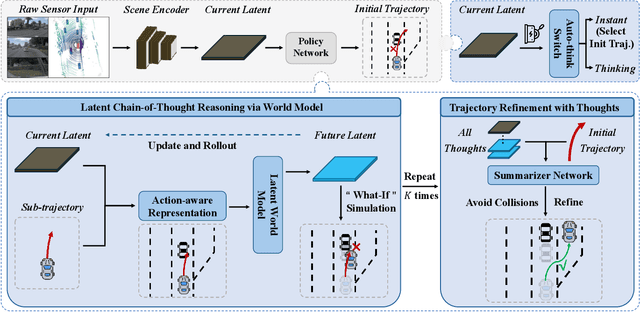
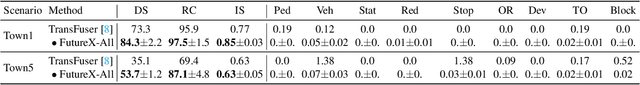
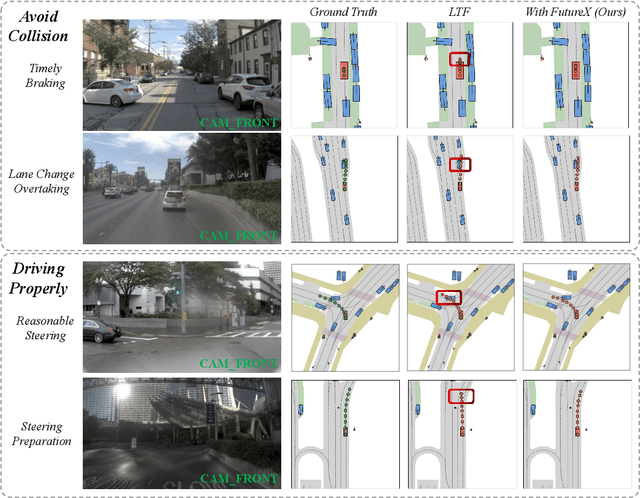
Abstract:In autonomous driving, end-to-end planners learn scene representations from raw sensor data and utilize them to generate a motion plan or control actions. However, exclusive reliance on the current scene for motion planning may result in suboptimal responses in highly dynamic traffic environments where ego actions further alter the future scene. To model the evolution of future scenes, we leverage the World Model to represent how the ego vehicle and its environment interact and change over time, which entails complex reasoning. The Chain of Thought (CoT) offers a promising solution by forecasting a sequence of future thoughts that subsequently guide trajectory refinement. In this paper, we propose FutureX, a CoT-driven pipeline that enhances end-to-end planners to perform complex motion planning via future scene latent reasoning and trajectory refinement. Specifically, the Auto-think Switch examines the current scene and decides whether additional reasoning is required to yield a higher-quality motion plan. Once FutureX enters the Thinking mode, the Latent World Model conducts a CoT-guided rollout to predict future scene representation, enabling the Summarizer Module to further refine the motion plan. Otherwise, FutureX operates in an Instant mode to generate motion plans in a forward pass for relatively simple scenes. Extensive experiments demonstrate that FutureX enhances existing methods by producing more rational motion plans and fewer collisions without compromising efficiency, thereby achieving substantial overall performance gains, e.g., 6.2 PDMS improvement for TransFuser on NAVSIM. Code will be released.
Carbon Price Forecasting with Structural Breaks: A Comparative Study of Deep Learning Models
Nov 07, 2025Abstract:Accurately forecasting carbon prices is essential for informed energy market decision-making, guiding sustainable energy planning, and supporting effective decarbonization strategies. However, it remains challenging due to structural breaks and high-frequency noise caused by frequent policy interventions and market shocks. Existing studies, including the most recent baseline approaches, have attempted to incorporate breakpoints but often treat denoising and modeling as separate processes and lack systematic evaluation across advanced deep learning architectures, limiting the robustness and the generalization capability. To address these gaps, this paper proposes a comprehensive hybrid framework that integrates structural break detection (Bai-Perron, ICSS, and PELT algorithms), wavelet signal denoising, and three state-of-the-art deep learning models (LSTM, GRU, and TCN). Using European Union Allowance (EUA) spot prices from 2007 to 2024 and exogenous features such as energy prices and policy indicators, the framework constructs univariate and multivariate datasets for comparative evaluation. Experimental results demonstrate that our proposed PELT-WT-TCN achieves the highest prediction accuracy, reducing forecasting errors by 22.35% in RMSE and 18.63% in MAE compared to the state-of-the-art baseline model (Breakpoints with Wavelet and LSTM), and by 70.55% in RMSE and 74.42% in MAE compared to the original LSTM without decomposition from the same baseline study. These findings underscore the value of integrating structural awareness and multiscale decomposition into deep learning architectures to enhance accuracy and interpretability in carbon price forecasting and other nonstationary financial time series.
Pathology-CoT: Learning Visual Chain-of-Thought Agent from Expert Whole Slide Image Diagnosis Behavior
Oct 06, 2025Abstract:Diagnosing a whole-slide image is an interactive, multi-stage process involving changes in magnification and movement between fields. Although recent pathology foundation models are strong, practical agentic systems that decide what field to examine next, adjust magnification, and deliver explainable diagnoses are still lacking. The blocker is data: scalable, clinically aligned supervision of expert viewing behavior that is tacit and experience-based, not written in textbooks or online, and therefore absent from large language model training. We introduce the AI Session Recorder, which works with standard WSI viewers to unobtrusively record routine navigation and convert the viewer logs into standardized behavioral commands (inspect or peek at discrete magnifications) and bounding boxes. A lightweight human-in-the-loop review turns AI-drafted rationales into the Pathology-CoT dataset, a form of paired "where to look" and "why it matters" supervision produced at roughly six times lower labeling time. Using this behavioral data, we build Pathologist-o3, a two-stage agent that first proposes regions of interest and then performs behavior-guided reasoning. On gastrointestinal lymph-node metastasis detection, it achieved 84.5% precision, 100.0% recall, and 75.4% accuracy, exceeding the state-of-the-art OpenAI o3 model and generalizing across backbones. To our knowledge, this constitutes one of the first behavior-grounded agentic systems in pathology. Turning everyday viewer logs into scalable, expert-validated supervision, our framework makes agentic pathology practical and establishes a path to human-aligned, upgradeable clinical AI.
FedAPM: Federated Learning via ADMM with Partial Model Personalization
Jun 05, 2025Abstract:In federated learning (FL), the assumption that datasets from different devices are independent and identically distributed (i.i.d.) often does not hold due to user differences, and the presence of various data modalities across clients makes using a single model impractical. Personalizing certain parts of the model can effectively address these issues by allowing those parts to differ across clients, while the remaining parts serve as a shared model. However, we found that partial model personalization may exacerbate client drift (each client's local model diverges from the shared model), thereby reducing the effectiveness and efficiency of FL algorithms. We propose an FL framework based on the alternating direction method of multipliers (ADMM), referred to as FedAPM, to mitigate client drift. We construct the augmented Lagrangian function by incorporating first-order and second-order proximal terms into the objective, with the second-order term providing fixed correction and the first-order term offering compensatory correction between the local and shared models. Our analysis demonstrates that FedAPM, by using explicit estimates of the Lagrange multiplier, is more stable and efficient in terms of convergence compared to other FL frameworks. We establish the global convergence of FedAPM training from arbitrary initial points to a stationary point, achieving three types of rates: constant, linear, and sublinear, under mild assumptions. We conduct experiments using four heterogeneous and multimodal datasets with different metrics to validate the performance of FedAPM. Specifically, FedAPM achieves faster and more accurate convergence, outperforming the SOTA methods with average improvements of 12.3% in test accuracy, 16.4% in F1 score, and 18.0% in AUC while requiring fewer communication rounds.
HAMF: A Hybrid Attention-Mamba Framework for Joint Scene Context Understanding and Future Motion Representation Learning
May 21, 2025



Abstract:Motion forecasting represents a critical challenge in autonomous driving systems, requiring accurate prediction of surrounding agents' future trajectories. While existing approaches predict future motion states with the extracted scene context feature from historical agent trajectories and road layouts, they suffer from the information degradation during the scene feature encoding. To address the limitation, we propose HAMF, a novel motion forecasting framework that learns future motion representations with the scene context encoding jointly, to coherently combine the scene understanding and future motion state prediction. We first embed the observed agent states and map information into 1D token sequences, together with the target multi-modal future motion features as a set of learnable tokens. Then we design a unified Attention-based encoder, which synergistically combines self-attention and cross-attention mechanisms to model the scene context information and aggregate future motion features jointly. Complementing the encoder, we implement the Mamba module in the decoding stage to further preserve the consistency and correlations among the learned future motion representations, to generate the accurate and diverse final trajectories. Extensive experiments on Argoverse 2 benchmark demonstrate that our hybrid Attention-Mamba model achieves state-of-the-art motion forecasting performance with the simple and lightweight architecture.
ReactDiff: Latent Diffusion for Facial Reaction Generation
May 20, 2025Abstract:Given the audio-visual clip of the speaker, facial reaction generation aims to predict the listener's facial reactions. The challenge lies in capturing the relevance between video and audio while balancing appropriateness, realism, and diversity. While prior works have mostly focused on uni-modal inputs or simplified reaction mappings, recent approaches such as PerFRDiff have explored multi-modal inputs and the one-to-many nature of appropriate reaction mappings. In this work, we propose the Facial Reaction Diffusion (ReactDiff) framework that uniquely integrates a Multi-Modality Transformer with conditional diffusion in the latent space for enhanced reaction generation. Unlike existing methods, ReactDiff leverages intra- and inter-class attention for fine-grained multi-modal interaction, while the latent diffusion process between the encoder and decoder enables diverse yet contextually appropriate outputs. Experimental results demonstrate that ReactDiff significantly outperforms existing approaches, achieving a facial reaction correlation of 0.26 and diversity score of 0.094 while maintaining competitive realism. The code is open-sourced at \href{https://github.com/Hunan-Tiger/ReactDiff}{github}.
UniCAD: Efficient and Extendable Architecture for Multi-Task Computer-Aided Diagnosis System
May 15, 2025Abstract:The growing complexity and scale of visual model pre-training have made developing and deploying multi-task computer-aided diagnosis (CAD) systems increasingly challenging and resource-intensive. Furthermore, the medical imaging community lacks an open-source CAD platform to enable the rapid creation of efficient and extendable diagnostic models. To address these issues, we propose UniCAD, a unified architecture that leverages the robust capabilities of pre-trained vision foundation models to seamlessly handle both 2D and 3D medical images while requiring only minimal task-specific parameters. UniCAD introduces two key innovations: (1) Efficiency: A low-rank adaptation strategy is employed to adapt a pre-trained visual model to the medical image domain, achieving performance on par with fully fine-tuned counterparts while introducing only 0.17% trainable parameters. (2) Plug-and-Play: A modular architecture that combines a frozen foundation model with multiple plug-and-play experts, enabling diverse tasks and seamless functionality expansion. Building on this unified CAD architecture, we establish an open-source platform where researchers can share and access lightweight CAD experts, fostering a more equitable and efficient research ecosystem. Comprehensive experiments across 12 diverse medical datasets demonstrate that UniCAD consistently outperforms existing methods in both accuracy and deployment efficiency. The source code and project page are available at https://mii-laboratory.github.io/UniCAD/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge