Dinggang Shen
Fellow, IEEE
S$^3$POT: Contrast-Driven Face Occlusion Segmentation via Self-Supervised Prompt Learning
Jan 31, 2026Abstract:Existing face parsing methods usually misclassify occlusions as facial components. This is because occlusion is a high-level concept, it does not refer to a concrete category of object. Thus, constructing a real-world face dataset covering all categories of occlusion object is almost impossible and accurate mask annotation is labor-intensive. To deal with the problems, we present S$^3$POT, a contrast-driven framework synergizing face generation with self-supervised spatial prompting, to achieve occlusion segmentation. The framework is inspired by the insights: 1) Modern face generators' ability to realistically reconstruct occluded regions, creating an image that preserve facial geometry while eliminating occlusion, and 2) Foundation segmentation models' (e.g., SAM) capacity to extract precise mask when provided with appropriate prompts. In particular, S$^3$POT consists of three modules: Reference Generation (RF), Feature enhancement (FE), and Prompt Selection (PS). First, a reference image is produced by RF using structural guidance from parsed mask. Second, FE performs contrast of tokens between raw and reference images to obtain an initial prompt, then modifies image features with the prompt by cross-attention. Third, based on the enhanced features, PS constructs a set of positive and negative prompts and screens them with a self-attention network for a mask decoder. The network is learned under the guidance of three novel and complementary objective functions without occlusion ground truth mask involved. Extensive experiments on a dedicatedly collected dataset demonstrate S$^3$POT's superior performance and the effectiveness of each module.
Adapting Foundation Model for Dental Caries Detection with Dual-View Co-Training
Aug 28, 2025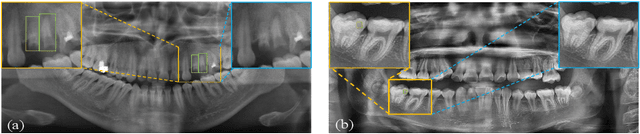
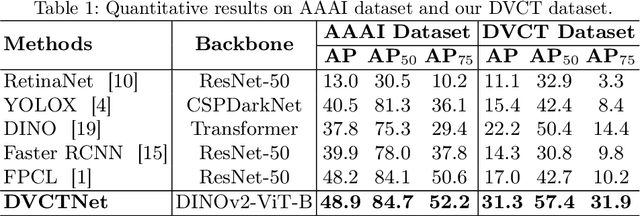
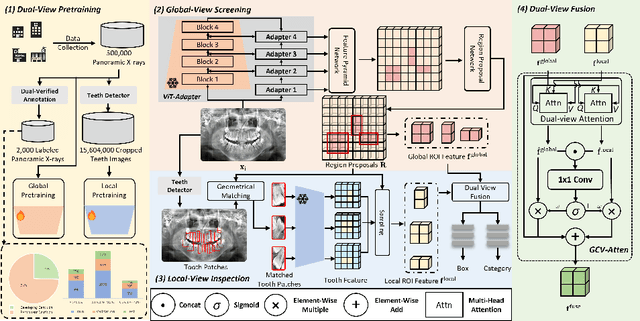
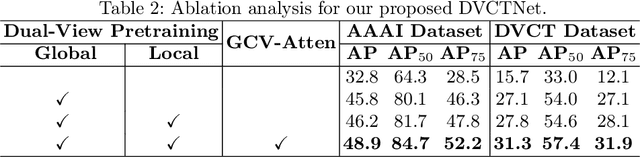
Abstract:Accurate dental caries detection from panoramic X-rays plays a pivotal role in preventing lesion progression. However, current detection methods often yield suboptimal accuracy due to subtle contrast variations and diverse lesion morphology of dental caries. In this work, inspired by the clinical workflow where dentists systematically combine whole-image screening with detailed tooth-level inspection, we present DVCTNet, a novel Dual-View Co-Training network for accurate dental caries detection. Our DVCTNet starts with employing automated tooth detection to establish two complementary views: a global view from panoramic X-ray images and a local view from cropped tooth images. We then pretrain two vision foundation models separately on the two views. The global-view foundation model serves as the detection backbone, generating region proposals and global features, while the local-view model extracts detailed features from corresponding cropped tooth patches matched by the region proposals. To effectively integrate information from both views, we introduce a Gated Cross-View Attention (GCV-Atten) module that dynamically fuses dual-view features, enhancing the detection pipeline by integrating the fused features back into the detection model for final caries detection. To rigorously evaluate our DVCTNet, we test it on a public dataset and further validate its performance on a newly curated, high-precision dental caries detection dataset, annotated using both intra-oral images and panoramic X-rays for double verification. Experimental results demonstrate DVCTNet's superior performance against existing state-of-the-art (SOTA) methods on both datasets, indicating the clinical applicability of our method. Our code and labeled dataset are available at https://github.com/ShanghaiTech-IMPACT/DVCTNet.
LGE-Guided Cross-Modality Contrastive Learning for Gadolinium-Free Cardiomyopathy Screening in Cine CMR
Aug 23, 2025Abstract:Cardiomyopathy, a principal contributor to heart failure and sudden cardiac mortality, demands precise early screening. Cardiac Magnetic Resonance (CMR), recognized as the diagnostic 'gold standard' through multiparametric protocols, holds the potential to serve as an accurate screening tool. However, its reliance on gadolinium contrast and labor-intensive interpretation hinders population-scale deployment. We propose CC-CMR, a Contrastive Learning and Cross-Modal alignment framework for gadolinium-free cardiomyopathy screening using cine CMR sequences. By aligning the latent spaces of cine CMR and Late Gadolinium Enhancement (LGE) sequences, our model encodes fibrosis-specific pathology into cine CMR embeddings. A Feature Interaction Module concurrently optimizes diagnostic precision and cross-modal feature congruence, augmented by an uncertainty-guided adaptive training mechanism that dynamically calibrates task-specific objectives to ensure model generalizability. Evaluated on multi-center data from 231 subjects, CC-CMR achieves accuracy of 0.943 (95% CI: 0.886-0.986), outperforming state-of-the-art cine-CMR-only models by 4.3% while eliminating gadolinium dependency, demonstrating its clinical viability for wide range of populations and healthcare environments.
Learning Interpretable Representations Leads to Semantically Faithful EEG-to-Text Generation
May 21, 2025Abstract:Pretrained generative models have opened new frontiers in brain decoding by enabling the synthesis of realistic texts and images from non-invasive brain recordings. However, the reliability of such outputs remains questionable--whether they truly reflect semantic activation in the brain, or are merely hallucinated by the powerful generative models. In this paper, we focus on EEG-to-text decoding and address its hallucination issue through the lens of posterior collapse. Acknowledging the underlying mismatch in information capacity between EEG and text, we reframe the decoding task as semantic summarization of core meanings rather than previously verbatim reconstruction of stimulus texts. To this end, we propose the Generative Language Inspection Model (GLIM), which emphasizes learning informative and interpretable EEG representations to improve semantic grounding under heterogeneous and small-scale data conditions. Experiments on the public ZuCo dataset demonstrate that GLIM consistently generates fluent, EEG-grounded sentences without teacher forcing. Moreover, it supports more robust evaluation beyond text similarity, through EEG-text retrieval and zero-shot semantic classification across sentiment categories, relation types, and corpus topics. Together, our architecture and evaluation protocols lay the foundation for reliable and scalable benchmarking in generative brain decoding.
UniCAD: Efficient and Extendable Architecture for Multi-Task Computer-Aided Diagnosis System
May 15, 2025Abstract:The growing complexity and scale of visual model pre-training have made developing and deploying multi-task computer-aided diagnosis (CAD) systems increasingly challenging and resource-intensive. Furthermore, the medical imaging community lacks an open-source CAD platform to enable the rapid creation of efficient and extendable diagnostic models. To address these issues, we propose UniCAD, a unified architecture that leverages the robust capabilities of pre-trained vision foundation models to seamlessly handle both 2D and 3D medical images while requiring only minimal task-specific parameters. UniCAD introduces two key innovations: (1) Efficiency: A low-rank adaptation strategy is employed to adapt a pre-trained visual model to the medical image domain, achieving performance on par with fully fine-tuned counterparts while introducing only 0.17% trainable parameters. (2) Plug-and-Play: A modular architecture that combines a frozen foundation model with multiple plug-and-play experts, enabling diverse tasks and seamless functionality expansion. Building on this unified CAD architecture, we establish an open-source platform where researchers can share and access lightweight CAD experts, fostering a more equitable and efficient research ecosystem. Comprehensive experiments across 12 diverse medical datasets demonstrate that UniCAD consistently outperforms existing methods in both accuracy and deployment efficiency. The source code and project page are available at https://mii-laboratory.github.io/UniCAD/.
A topology-preserving three-stage framework for fully-connected coronary artery extraction
Apr 02, 2025Abstract:Coronary artery extraction is a crucial prerequisite for computer-aided diagnosis of coronary artery disease. Accurately extracting the complete coronary tree remains challenging due to several factors, including presence of thin distal vessels, tortuous topological structures, and insufficient contrast. These issues often result in over-segmentation and under-segmentation in current segmentation methods. To address these challenges, we propose a topology-preserving three-stage framework for fully-connected coronary artery extraction. This framework includes vessel segmentation, centerline reconnection, and missing vessel reconstruction. First, we introduce a new centerline enhanced loss in the segmentation process. Second, for the broken vessel segments, we further propose a regularized walk algorithm to integrate distance, probabilities predicted by a centerline classifier, and directional cosine similarity, for reconnecting the centerlines. Third, we apply implicit neural representation and implicit modeling, to reconstruct the geometric model of the missing vessels. Experimental results show that our proposed framework outperforms existing methods, achieving Dice scores of 88.53\% and 85.07\%, with Hausdorff Distances (HD) of 1.07mm and 1.63mm on ASOCA and PDSCA datasets, respectively. Code will be available at https://github.com/YH-Qiu/CorSegRec.
MAST-Pro: Dynamic Mixture-of-Experts for Adaptive Segmentation of Pan-Tumors with Knowledge-Driven Prompts
Mar 18, 2025Abstract:Accurate tumor segmentation is crucial for cancer diagnosis and treatment. While foundation models have advanced general-purpose segmentation, existing methods still struggle with: (1) limited incorporation of medical priors, (2) imbalance between generic and tumor-specific features, and (3) high computational costs for clinical adaptation. To address these challenges, we propose MAST-Pro (Mixture-of-experts for Adaptive Segmentation of pan-Tumors with knowledge-driven Prompts), a novel framework that integrates dynamic Mixture-of-Experts (D-MoE) and knowledge-driven prompts for pan-tumor segmentation. Specifically, text and anatomical prompts provide domain-specific priors, guiding tumor representation learning, while D-MoE dynamically selects experts to balance generic and tumor-specific feature learning, improving segmentation accuracy across diverse tumor types. To enhance efficiency, we employ Parameter-Efficient Fine-Tuning (PEFT), optimizing MAST-Pro with significantly reduced computational overhead. Experiments on multi-anatomical tumor datasets demonstrate that MAST-Pro outperforms state-of-the-art approaches, achieving up to a 5.20% improvement in average DSC while reducing trainable parameters by 91.04%, without compromising accuracy.
3D MedDiffusion: A 3D Medical Diffusion Model for Controllable and High-quality Medical Image Generation
Dec 17, 2024Abstract:The generation of medical images presents significant challenges due to their high-resolution and three-dimensional nature. Existing methods often yield suboptimal performance in generating high-quality 3D medical images, and there is currently no universal generative framework for medical imaging. In this paper, we introduce the 3D Medical Diffusion (3D MedDiffusion) model for controllable, high-quality 3D medical image generation. 3D MedDiffusion incorporates a novel, highly efficient Patch-Volume Autoencoder that compresses medical images into latent space through patch-wise encoding and recovers back into image space through volume-wise decoding. Additionally, we design a new noise estimator to capture both local details and global structure information during diffusion denoising process. 3D MedDiffusion can generate fine-detailed, high-resolution images (up to 512x512x512) and effectively adapt to various downstream tasks as it is trained on large-scale datasets covering CT and MRI modalities and different anatomical regions (from head to leg). Experimental results demonstrate that 3D MedDiffusion surpasses state-of-the-art methods in generative quality and exhibits strong generalizability across tasks such as sparse-view CT reconstruction, fast MRI reconstruction, and data augmentation.
End-to-end Triple-domain PET Enhancement: A Hybrid Denoising-and-reconstruction Framework for Reconstructing Standard-dose PET Images from Low-dose PET Sinograms
Dec 04, 2024
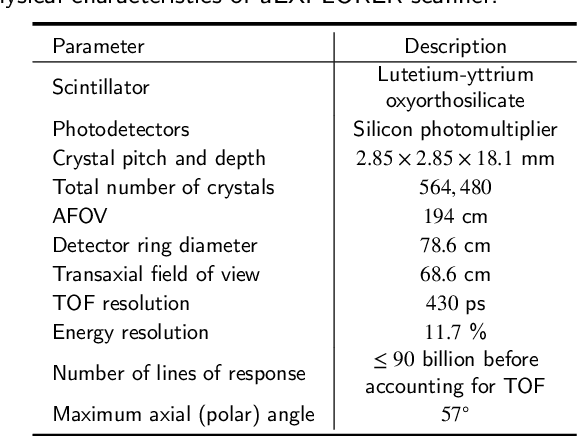

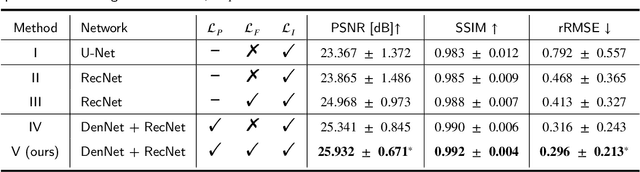
Abstract:As a sensitive functional imaging technique, positron emission tomography (PET) plays a critical role in early disease diagnosis. However, obtaining a high-quality PET image requires injecting a sufficient dose (standard dose) of radionuclides into the body, which inevitably poses radiation hazards to patients. To mitigate radiation hazards, the reconstruction of standard-dose PET (SPET) from low-dose PET (LPET) is desired. According to imaging theory, PET reconstruction process involves multiple domains (e.g., projection domain and image domain), and a significant portion of the difference between SPET and LPET arises from variations in the noise levels introduced during the sampling of raw data as sinograms. In light of these two facts, we propose an end-to-end TriPle-domain LPET EnhancemenT (TriPLET) framework, by leveraging the advantages of a hybrid denoising-and-reconstruction process and a triple-domain representation (i.e., sinograms, frequency spectrum maps, and images) to reconstruct SPET images from LPET sinograms. Specifically, TriPLET consists of three sequentially coupled components including 1) a Transformer-assisted denoising network that denoises the inputted LPET sinograms in the projection domain, 2) a discrete-wavelet-transform-based reconstruction network that further reconstructs SPET from LPET in the wavelet domain, and 3) a pair-based adversarial network that evaluates the reconstructed SPET images in the image domain. Extensive experiments on the real PET dataset demonstrate that our proposed TriPLET can reconstruct SPET images with the highest similarity and signal-to-noise ratio to real data, compared with state-of-the-art methods.
DVasMesh: Deep Structured Mesh Reconstruction from Vascular Images for Dynamics Modeling of Vessels
Dec 01, 2024



Abstract:Vessel dynamics simulation is vital in studying the relationship between geometry and vascular disease progression. Reliable dynamics simulation relies on high-quality vascular meshes. Most of the existing mesh generation methods highly depend on manual annotation, which is time-consuming and laborious, usually facing challenges such as branch merging and vessel disconnection. This will hinder vessel dynamics simulation, especially for the population study. To address this issue, we propose a deep learning-based method, dubbed as DVasMesh to directly generate structured hexahedral vascular meshes from vascular images. Our contributions are threefold. First, we propose to formally formulate each vertex of the vascular graph by a four-element vector, including coordinates of the centerline point and the radius. Second, a vectorized graph template is employed to guide DVasMesh to estimate the vascular graph. Specifically, we introduce a sampling operator, which samples the extracted features of the vascular image (by a segmentation network) according to the vertices in the template graph. Third, we employ a graph convolution network (GCN) and take the sampled features as nodes to estimate the deformation between vertices of the template graph and target graph, and the deformed graph template is used to build the mesh. Taking advantage of end-to-end learning and discarding direct dependency on annotated labels, our DVasMesh demonstrates outstanding performance in generating structured vascular meshes on cardiac and cerebral vascular images. It shows great potential for clinical applications by reducing mesh generation time from 2 hours (manual) to 30 seconds (automatic).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge