Dengqiang Jia
DVasMesh: Deep Structured Mesh Reconstruction from Vascular Images for Dynamics Modeling of Vessels
Dec 01, 2024



Abstract:Vessel dynamics simulation is vital in studying the relationship between geometry and vascular disease progression. Reliable dynamics simulation relies on high-quality vascular meshes. Most of the existing mesh generation methods highly depend on manual annotation, which is time-consuming and laborious, usually facing challenges such as branch merging and vessel disconnection. This will hinder vessel dynamics simulation, especially for the population study. To address this issue, we propose a deep learning-based method, dubbed as DVasMesh to directly generate structured hexahedral vascular meshes from vascular images. Our contributions are threefold. First, we propose to formally formulate each vertex of the vascular graph by a four-element vector, including coordinates of the centerline point and the radius. Second, a vectorized graph template is employed to guide DVasMesh to estimate the vascular graph. Specifically, we introduce a sampling operator, which samples the extracted features of the vascular image (by a segmentation network) according to the vertices in the template graph. Third, we employ a graph convolution network (GCN) and take the sampled features as nodes to estimate the deformation between vertices of the template graph and target graph, and the deformed graph template is used to build the mesh. Taking advantage of end-to-end learning and discarding direct dependency on annotated labels, our DVasMesh demonstrates outstanding performance in generating structured vascular meshes on cardiac and cerebral vascular images. It shows great potential for clinical applications by reducing mesh generation time from 2 hours (manual) to 30 seconds (automatic).
Unleashing the Strengths of Unlabeled Data in Pan-cancer Abdominal Organ Quantification: the FLARE22 Challenge
Aug 10, 2023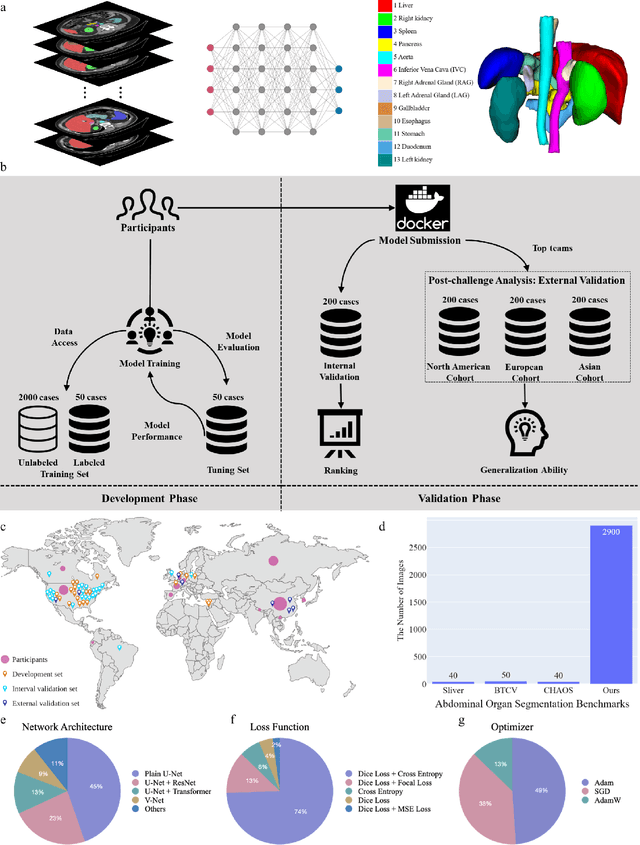
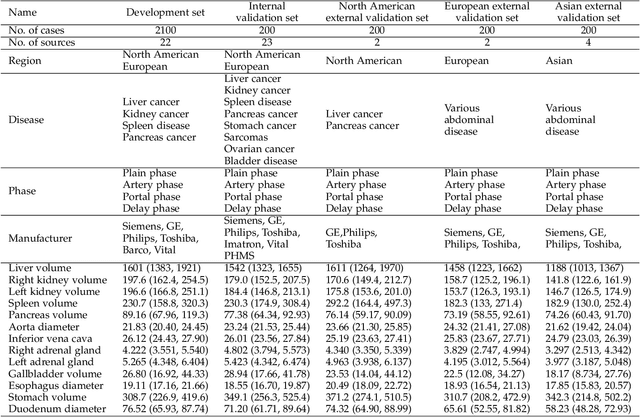
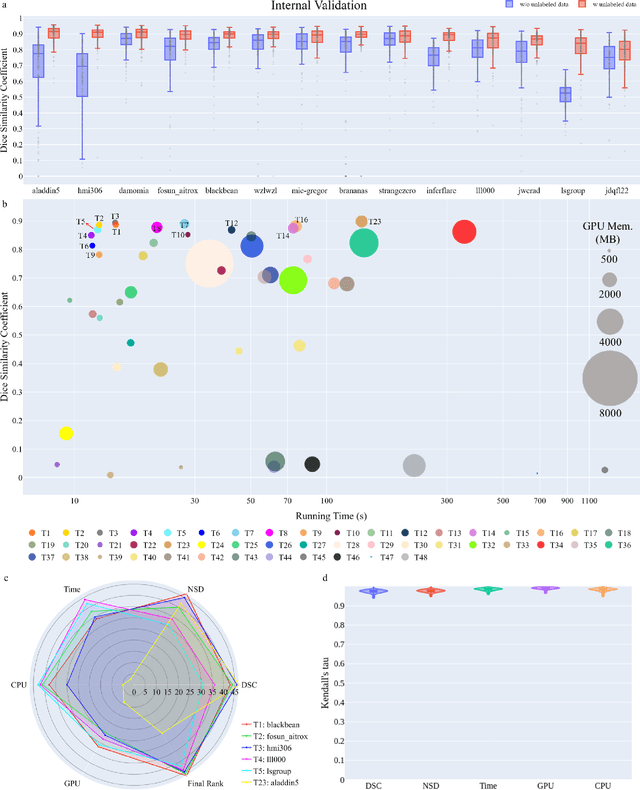
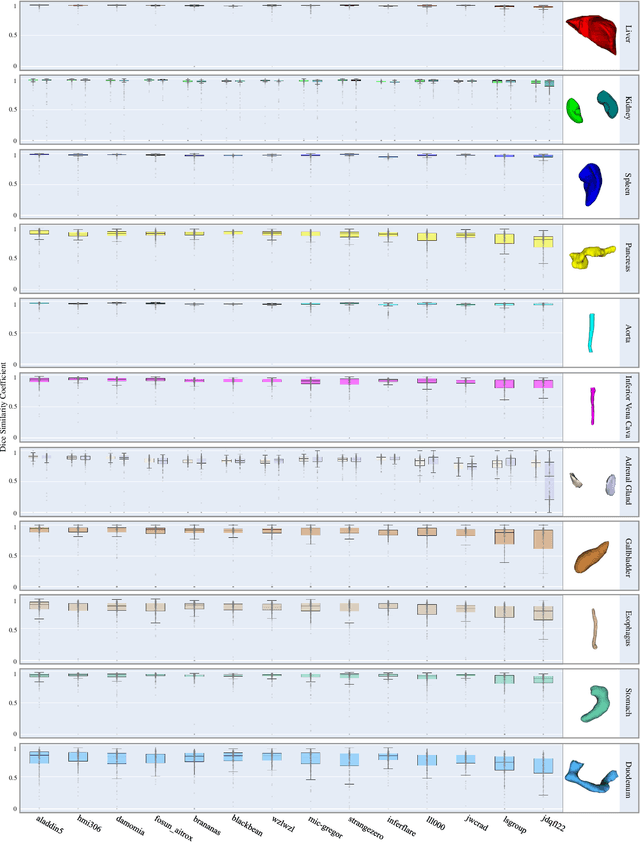
Abstract:Quantitative organ assessment is an essential step in automated abdominal disease diagnosis and treatment planning. Artificial intelligence (AI) has shown great potential to automatize this process. However, most existing AI algorithms rely on many expert annotations and lack a comprehensive evaluation of accuracy and efficiency in real-world multinational settings. To overcome these limitations, we organized the FLARE 2022 Challenge, the largest abdominal organ analysis challenge to date, to benchmark fast, low-resource, accurate, annotation-efficient, and generalized AI algorithms. We constructed an intercontinental and multinational dataset from more than 50 medical groups, including Computed Tomography (CT) scans with different races, diseases, phases, and manufacturers. We independently validated that a set of AI algorithms achieved a median Dice Similarity Coefficient (DSC) of 90.0\% by using 50 labeled scans and 2000 unlabeled scans, which can significantly reduce annotation requirements. The best-performing algorithms successfully generalized to holdout external validation sets, achieving a median DSC of 89.5\%, 90.9\%, and 88.3\% on North American, European, and Asian cohorts, respectively. They also enabled automatic extraction of key organ biology features, which was labor-intensive with traditional manual measurements. This opens the potential to use unlabeled data to boost performance and alleviate annotation shortages for modern AI models.
Multi-Target Landmark Detection with Incomplete Images via Reinforcement Learning and Shape Prior
Jan 13, 2023



Abstract:Medical images are generally acquired with limited field-of-view (FOV), which could lead to incomplete regions of interest (ROI), and thus impose a great challenge on medical image analysis. This is particularly evident for the learning-based multi-target landmark detection, where algorithms could be misleading to learn primarily the variation of background due to the varying FOV, failing the detection of targets. Based on learning a navigation policy, instead of predicting targets directly, reinforcement learning (RL)-based methods have the potential totackle this challenge in an efficient manner. Inspired by this, in this work we propose a multi-agent RL framework for simultaneous multi-target landmark detection. This framework is aimed to learn from incomplete or (and) complete images to form an implicit knowledge of global structure, which is consolidated during the training stage for the detection of targets from either complete or incomplete test images. To further explicitly exploit the global structural information from incomplete images, we propose to embed a shape model into the RL process. With this prior knowledge, the proposed RL model can not only localize dozens of targetssimultaneously, but also work effectively and robustly in the presence of incomplete images. We validated the applicability and efficacy of the proposed method on various multi-target detection tasks with incomplete images from practical clinics, using body dual-energy X-ray absorptiometry (DXA), cardiac MRI and head CT datasets. Results showed that our method could predict whole set of landmarks with incomplete training images up to 80% missing proportion (average distance error 2.29 cm on body DXA), and could detect unseen landmarks in regions with missing image information outside FOV of target images (average distance error 6.84 mm on 3D half-head CT).
A low-rank representation for unsupervised registration of medical images
May 20, 2021



Abstract:Registration networks have shown great application potentials in medical image analysis. However, supervised training methods have a great demand for large and high-quality labeled datasets, which is time-consuming and sometimes impractical due to data sharing issues. Unsupervised image registration algorithms commonly employ intensity-based similarity measures as loss functions without any manual annotations. These methods estimate the parameterized transformations between pairs of moving and fixed images through the optimization of the network parameters during training. However, these methods become less effective when the image quality varies, e.g., some images are corrupted by substantial noise or artifacts. In this work, we propose a novel approach based on a low-rank representation, i.e., Regnet-LRR, to tackle the problem. We project noisy images into a noise-free low-rank space, and then compute the similarity between the images. Based on the low-rank similarity measure, we train the registration network to predict the dense deformation fields of noisy image pairs. We highlight that the low-rank projection is reformulated in a way that the registration network can successfully update gradients. With two tasks, i.e., cardiac and abdominal intra-modality registration, we demonstrate that the low-rank representation can boost the generalization ability and robustness of models as well as bring significant improvements in noisy data registration scenarios.
Learning-Based Algorithms for Vessel Tracking: A Review
Dec 16, 2020



Abstract:Developing efficient vessel-tracking algorithms is crucial for imaging-based diagnosis and treatment of vascular diseases. Vessel tracking aims to solve recognition problems such as key (seed) point detection, centerline extraction, and vascular segmentation. Extensive image-processing techniques have been developed to overcome the problems of vessel tracking that are mainly attributed to the complex morphologies of vessels and image characteristics of angiography. This paper presents a literature review on vessel-tracking methods, focusing on machine-learning-based methods. First, the conventional machine-learning-based algorithms are reviewed, and then, a general survey of deep-learning-based frameworks is provided. On the basis of the reviewed methods, the evaluation issues are introduced. The paper is concluded with discussions about the remaining exigencies and future research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge