Shangqi Gao
R$^{2}$Seg: Training-Free OOD Medical Tumor Segmentation via Anatomical Reasoning and Statistical Rejection
Nov 16, 2025Abstract:Foundation models for medical image segmentation struggle under out-of-distribution (OOD) shifts, often producing fragmented false positives on OOD tumors. We introduce R$^{2}$Seg, a training-free framework for robust OOD tumor segmentation that operates via a two-stage Reason-and-Reject process. First, the Reason step employs an LLM-guided anatomical reasoning planner to localize organ anchors and generate multi-scale ROIs. Second, the Reject step applies two-sample statistical testing to candidates generated by a frozen foundation model (BiomedParse) within these ROIs. This statistical rejection filter retains only candidates significantly different from normal tissue, effectively suppressing false positives. Our framework requires no parameter updates, making it compatible with zero-update test-time augmentation and avoiding catastrophic forgetting. On multi-center and multi-modal tumor segmentation benchmarks, R$^{2}$Seg substantially improves Dice, specificity, and sensitivity over strong baselines and the original foundation models. Code are available at https://github.com/Eurekashen/R2Seg.
Integrating Pathology and CT Imaging for Personalized Recurrence Risk Prediction in Renal Cancer
Aug 29, 2025Abstract:Recurrence risk estimation in clear cell renal cell carcinoma (ccRCC) is essential for guiding postoperative surveillance and treatment. The Leibovich score remains widely used for stratifying distant recurrence risk but offers limited patient-level resolution and excludes imaging information. This study evaluates multimodal recurrence prediction by integrating preoperative computed tomography (CT) and postoperative histopathology whole-slide images (WSIs). A modular deep learning framework with pretrained encoders and Cox-based survival modeling was tested across unimodal, late fusion, and intermediate fusion setups. In a real-world ccRCC cohort, WSI-based models consistently outperformed CT-only models, underscoring the prognostic strength of pathology. Intermediate fusion further improved performance, with the best model (TITAN-CONCH with ResNet-18) approaching the adjusted Leibovich score. Random tie-breaking narrowed the gap between the clinical baseline and learned models, suggesting discretization may overstate individualized performance. Using simple embedding concatenation, radiology added value primarily through fusion. These findings demonstrate the feasibility of foundation model-based multimodal integration for personalized ccRCC risk prediction. Future work should explore more expressive fusion strategies, larger multimodal datasets, and general-purpose CT encoders to better match pathology modeling capacity.
Learning Concept-Driven Logical Rules for Interpretable and Generalizable Medical Image Classification
May 20, 2025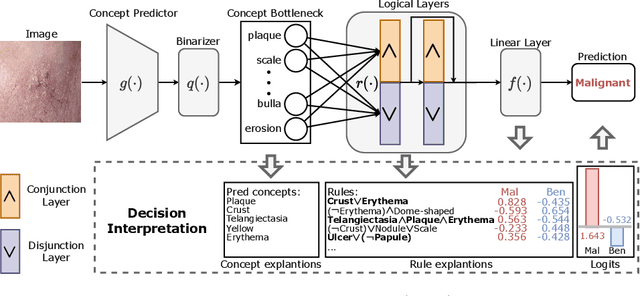
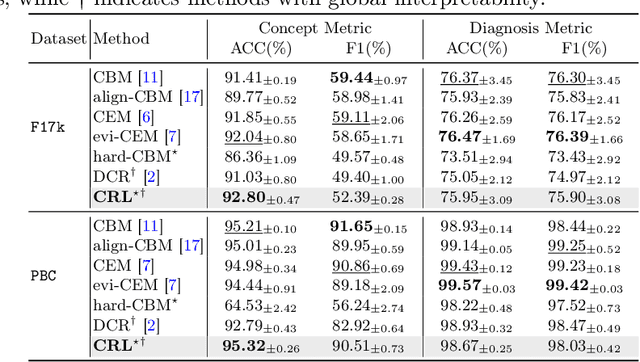
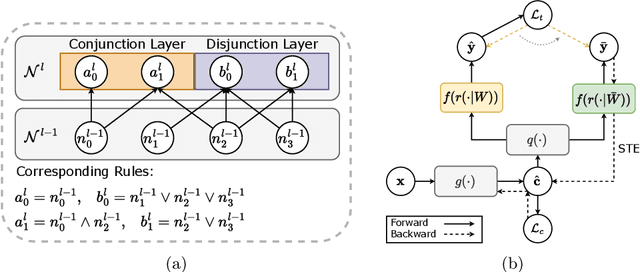
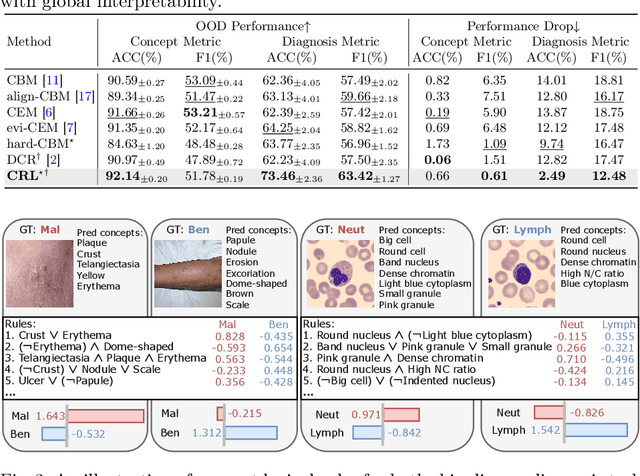
Abstract:The pursuit of decision safety in clinical applications highlights the potential of concept-based methods in medical imaging. While these models offer active interpretability, they often suffer from concept leakages, where unintended information within soft concept representations undermines both interpretability and generalizability. Moreover, most concept-based models focus solely on local explanations (instance-level), neglecting the global decision logic (dataset-level). To address these limitations, we propose Concept Rule Learner (CRL), a novel framework to learn Boolean logical rules from binarized visual concepts. CRL employs logical layers to capture concept correlations and extract clinically meaningful rules, thereby providing both local and global interpretability. Experiments on two medical image classification tasks show that CRL achieves competitive performance with existing methods while significantly improving generalizability to out-of-distribution data. The code of our work is available at https://github.com/obiyoag/crl.
Empowering Medical Multi-Agents with Clinical Consultation Flow for Dynamic Diagnosis
Mar 19, 2025



Abstract:Traditional AI-based healthcare systems often rely on single-modal data, limiting diagnostic accuracy due to incomplete information. However, recent advancements in foundation models show promising potential for enhancing diagnosis combining multi-modal information. While these models excel in static tasks, they struggle with dynamic diagnosis, failing to manage multi-turn interactions and often making premature diagnostic decisions due to insufficient persistence in information collection.To address this, we propose a multi-agent framework inspired by consultation flow and reinforcement learning (RL) to simulate the entire consultation process, integrating multiple clinical information for effective diagnosis. Our approach incorporates a hierarchical action set, structured from clinic consultation flow and medical textbook, to effectively guide the decision-making process. This strategy improves agent interactions, enabling them to adapt and optimize actions based on the dynamic state. We evaluated our framework on a public dynamic diagnosis benchmark. The proposed framework evidentially improves the baseline methods and achieves state-of-the-art performance compared to existing foundation model-based methods.
InDeed: Interpretable image deep decomposition with guaranteed generalizability
Jan 02, 2025



Abstract:Image decomposition aims to analyze an image into elementary components, which is essential for numerous downstream tasks and also by nature provides certain interpretability to the analysis. Deep learning can be powerful for such tasks, but surprisingly their combination with a focus on interpretability and generalizability is rarely explored. In this work, we introduce a novel framework for interpretable deep image decomposition, combining hierarchical Bayesian modeling and deep learning to create an architecture-modularized and model-generalizable deep neural network (DNN). The proposed framework includes three steps: (1) hierarchical Bayesian modeling of image decomposition, (2) transforming the inference problem into optimization tasks, and (3) deep inference via a modularized Bayesian DNN. We further establish a theoretical connection between the loss function and the generalization error bound, which inspires a new test-time adaptation approach for out-of-distribution scenarios. We instantiated the application using two downstream tasks, \textit{i.e.}, image denoising, and unsupervised anomaly detection, and the results demonstrated improved generalizability as well as interpretability of our methods. The source code will be released upon the acceptance of this paper.
Trustworthy Contrast-enhanced Brain MRI Synthesis
Jul 10, 2024



Abstract:Contrast-enhanced brain MRI (CE-MRI) is a valuable diagnostic technique but may pose health risks and incur high costs. To create safer alternatives, multi-modality medical image translation aims to synthesize CE-MRI images from other available modalities. Although existing methods can generate promising predictions, they still face two challenges, i.e., exhibiting over-confidence and lacking interpretability on predictions. To address the above challenges, this paper introduces TrustI2I, a novel trustworthy method that reformulates multi-to-one medical image translation problem as a multimodal regression problem, aiming to build an uncertainty-aware and reliable system. Specifically, our method leverages deep evidential regression to estimate prediction uncertainties and employs an explicit intermediate and late fusion strategy based on the Mixture of Normal Inverse Gamma (MoNIG) distribution, enhancing both synthesis quality and interpretability. Additionally, we incorporate uncertainty calibration to improve the reliability of uncertainty. Validation on the BraTS2018 dataset demonstrates that our approach surpasses current methods, producing higher-quality images with rational uncertainty estimation.
Multi-Center Fetal Brain Tissue Annotation (FeTA) Challenge 2022 Results
Feb 08, 2024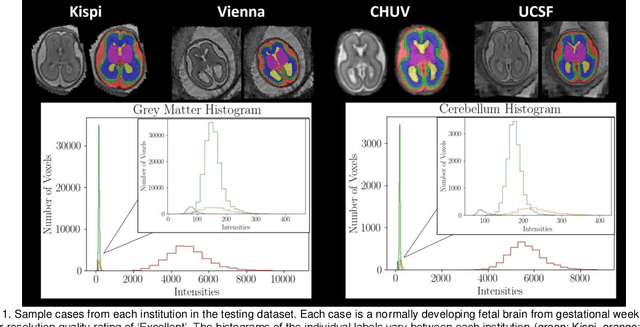
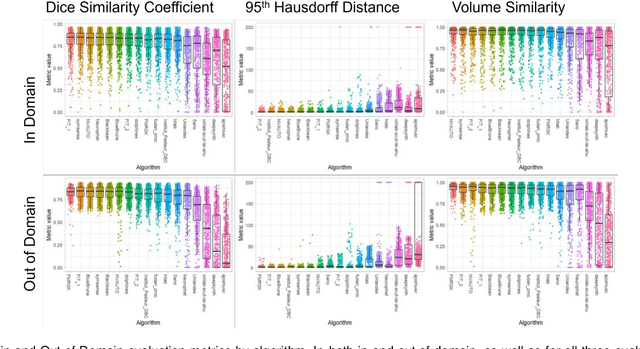
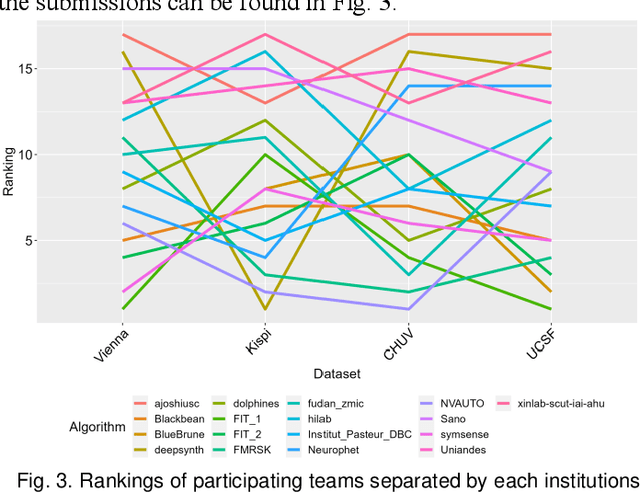
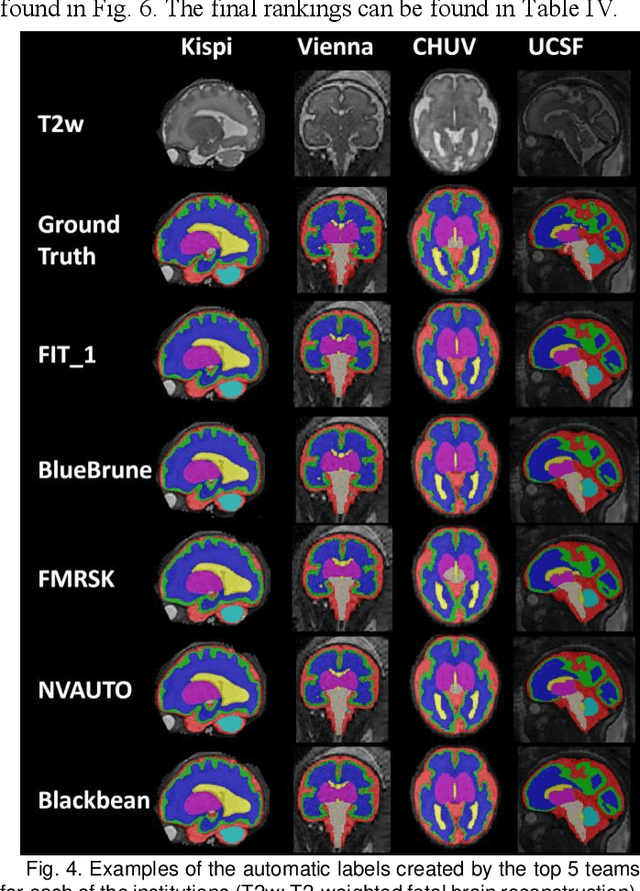
Abstract:Segmentation is a critical step in analyzing the developing human fetal brain. There have been vast improvements in automatic segmentation methods in the past several years, and the Fetal Brain Tissue Annotation (FeTA) Challenge 2021 helped to establish an excellent standard of fetal brain segmentation. However, FeTA 2021 was a single center study, and the generalizability of algorithms across different imaging centers remains unsolved, limiting real-world clinical applicability. The multi-center FeTA Challenge 2022 focuses on advancing the generalizability of fetal brain segmentation algorithms for magnetic resonance imaging (MRI). In FeTA 2022, the training dataset contained images and corresponding manually annotated multi-class labels from two imaging centers, and the testing data contained images from these two imaging centers as well as two additional unseen centers. The data from different centers varied in many aspects, including scanners used, imaging parameters, and fetal brain super-resolution algorithms applied. 16 teams participated in the challenge, and 17 algorithms were evaluated. Here, a detailed overview and analysis of the challenge results are provided, focusing on the generalizability of the submissions. Both in- and out of domain, the white matter and ventricles were segmented with the highest accuracy, while the most challenging structure remains the cerebral cortex due to anatomical complexity. The FeTA Challenge 2022 was able to successfully evaluate and advance generalizability of multi-class fetal brain tissue segmentation algorithms for MRI and it continues to benchmark new algorithms. The resulting new methods contribute to improving the analysis of brain development in utero.
BayeSeg: Bayesian Modeling for Medical Image Segmentation with Interpretable Generalizability
Mar 03, 2023Abstract:Due to the cross-domain distribution shift aroused from diverse medical imaging systems, many deep learning segmentation methods fail to perform well on unseen data, which limits their real-world applicability. Recent works have shown the benefits of extracting domain-invariant representations on domain generalization. However, the interpretability of domain-invariant features remains a great challenge. To address this problem, we propose an interpretable Bayesian framework (BayeSeg) through Bayesian modeling of image and label statistics to enhance model generalizability for medical image segmentation. Specifically, we first decompose an image into a spatial-correlated variable and a spatial-variant variable, assigning hierarchical Bayesian priors to explicitly force them to model the domain-stable shape and domain-specific appearance information respectively. Then, we model the segmentation as a locally smooth variable only related to the shape. Finally, we develop a variational Bayesian framework to infer the posterior distributions of these explainable variables. The framework is implemented with neural networks, and thus is referred to as deep Bayesian segmentation. Quantitative and qualitative experimental results on prostate segmentation and cardiac segmentation tasks have shown the effectiveness of our proposed method. Moreover, we investigated the interpretability of BayeSeg by explaining the posteriors and analyzed certain factors that affect the generalization ability through further ablation studies. Our code will be released via https://zmiclab.github.io/projects.html, once the manuscript is accepted for publication.
Multi-Target Landmark Detection with Incomplete Images via Reinforcement Learning and Shape Prior
Jan 13, 2023



Abstract:Medical images are generally acquired with limited field-of-view (FOV), which could lead to incomplete regions of interest (ROI), and thus impose a great challenge on medical image analysis. This is particularly evident for the learning-based multi-target landmark detection, where algorithms could be misleading to learn primarily the variation of background due to the varying FOV, failing the detection of targets. Based on learning a navigation policy, instead of predicting targets directly, reinforcement learning (RL)-based methods have the potential totackle this challenge in an efficient manner. Inspired by this, in this work we propose a multi-agent RL framework for simultaneous multi-target landmark detection. This framework is aimed to learn from incomplete or (and) complete images to form an implicit knowledge of global structure, which is consolidated during the training stage for the detection of targets from either complete or incomplete test images. To further explicitly exploit the global structural information from incomplete images, we propose to embed a shape model into the RL process. With this prior knowledge, the proposed RL model can not only localize dozens of targetssimultaneously, but also work effectively and robustly in the presence of incomplete images. We validated the applicability and efficacy of the proposed method on various multi-target detection tasks with incomplete images from practical clinics, using body dual-energy X-ray absorptiometry (DXA), cardiac MRI and head CT datasets. Results showed that our method could predict whole set of landmarks with incomplete training images up to 80% missing proportion (average distance error 2.29 cm on body DXA), and could detect unseen landmarks in regions with missing image information outside FOV of target images (average distance error 6.84 mm on 3D half-head CT).
Joint Modeling of Image and Label Statistics for Enhancing Model Generalizability of Medical Image Segmentation
Jun 09, 2022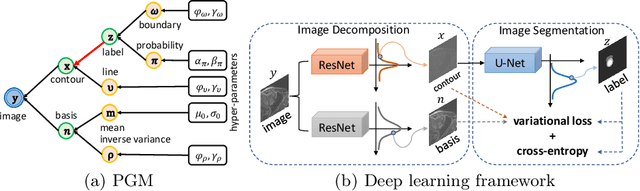
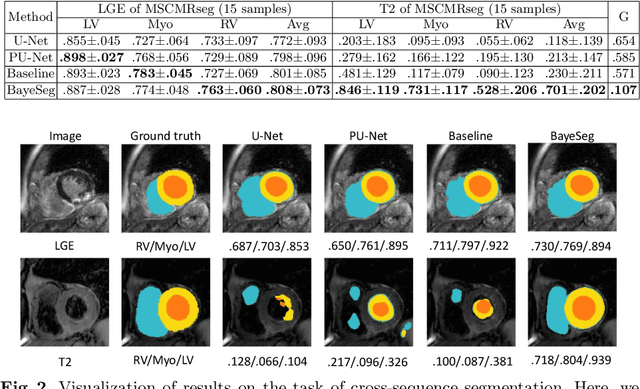
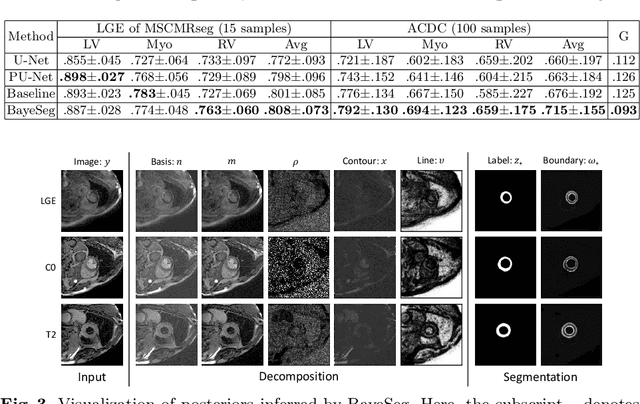
Abstract:Although supervised deep-learning has achieved promising performance in medical image segmentation, many methods cannot generalize well on unseen data, limiting their real-world applicability. To address this problem, we propose a deep learning-based Bayesian framework, which jointly models image and label statistics, utilizing the domain-irrelevant contour of a medical image for segmentation. Specifically, we first decompose an image into components of contour and basis. Then, we model the expected label as a variable only related to the contour. Finally, we develop a variational Bayesian framework to infer the posterior distributions of these variables, including the contour, the basis, and the label. The framework is implemented with neural networks, thus is referred to as deep Bayesian segmentation. Results on the task of cross-sequence cardiac MRI segmentation show that our method set a new state of the art for model generalizability. Particularly, the BayeSeg model trained with LGE MRI generalized well on T2 images and outperformed other models with great margins, i.e., over 0.47 in terms of average Dice. Our code is available at https://zmiclab.github.io/projects.html.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge