Priscille de Dumast
Department of Radiology, Lausanne University Hospital, CIBM Center for Biomedical Imaging, Switzerland
Multi-Center Fetal Brain Tissue Annotation (FeTA) Challenge 2022 Results
Feb 08, 2024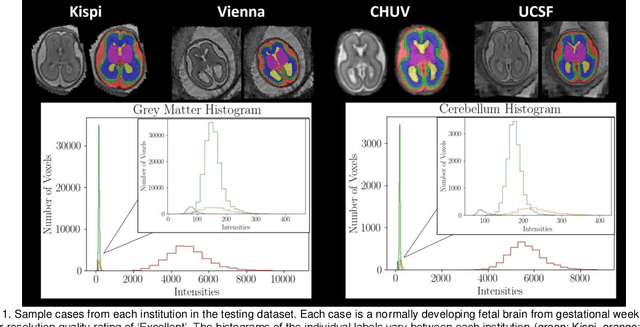
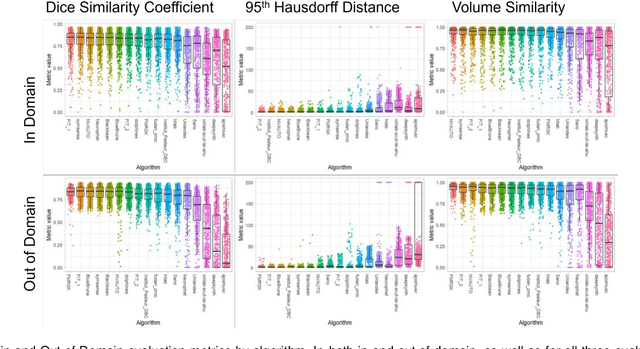
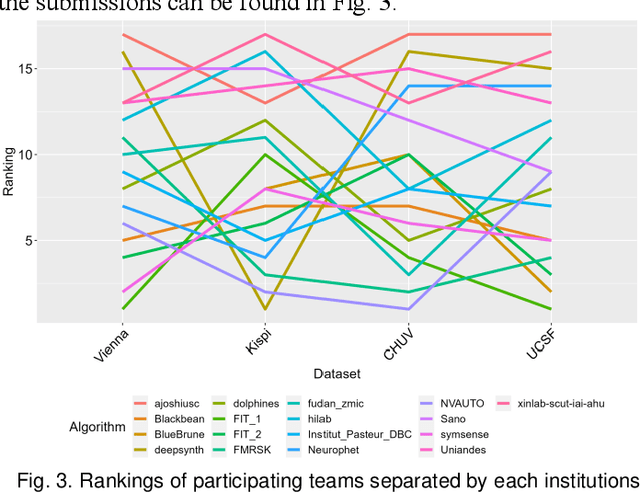
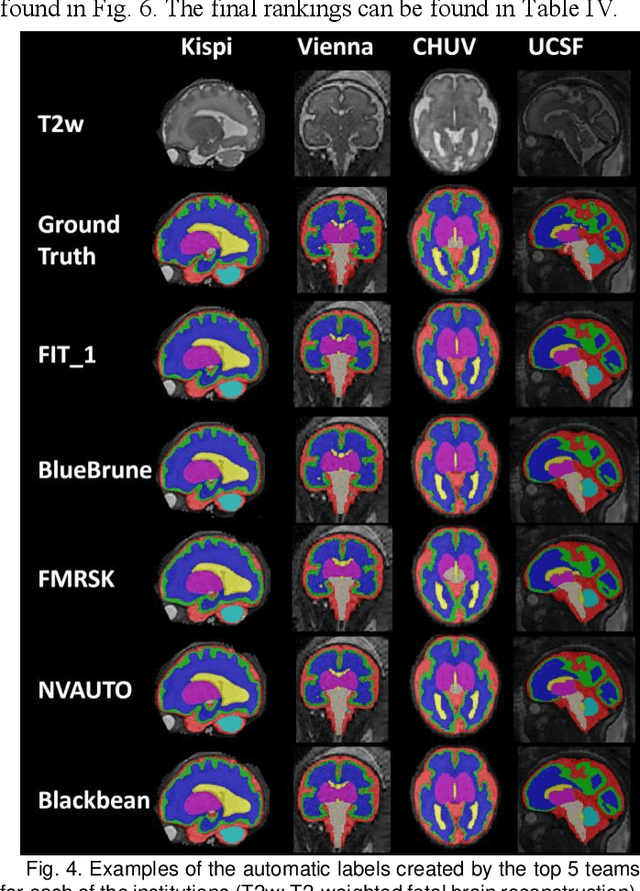
Abstract:Segmentation is a critical step in analyzing the developing human fetal brain. There have been vast improvements in automatic segmentation methods in the past several years, and the Fetal Brain Tissue Annotation (FeTA) Challenge 2021 helped to establish an excellent standard of fetal brain segmentation. However, FeTA 2021 was a single center study, and the generalizability of algorithms across different imaging centers remains unsolved, limiting real-world clinical applicability. The multi-center FeTA Challenge 2022 focuses on advancing the generalizability of fetal brain segmentation algorithms for magnetic resonance imaging (MRI). In FeTA 2022, the training dataset contained images and corresponding manually annotated multi-class labels from two imaging centers, and the testing data contained images from these two imaging centers as well as two additional unseen centers. The data from different centers varied in many aspects, including scanners used, imaging parameters, and fetal brain super-resolution algorithms applied. 16 teams participated in the challenge, and 17 algorithms were evaluated. Here, a detailed overview and analysis of the challenge results are provided, focusing on the generalizability of the submissions. Both in- and out of domain, the white matter and ventricles were segmented with the highest accuracy, while the most challenging structure remains the cerebral cortex due to anatomical complexity. The FeTA Challenge 2022 was able to successfully evaluate and advance generalizability of multi-class fetal brain tissue segmentation algorithms for MRI and it continues to benchmark new algorithms. The resulting new methods contribute to improving the analysis of brain development in utero.
Domain generalization in fetal brain MRI segmentation \\with multi-reconstruction augmentation
Nov 25, 2022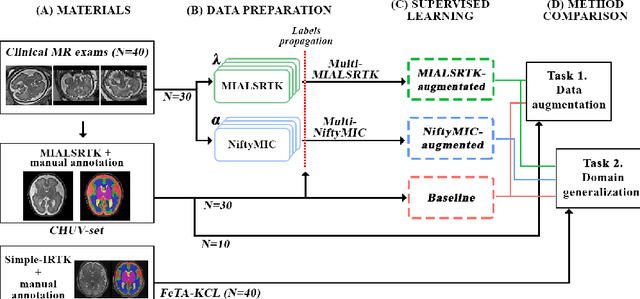

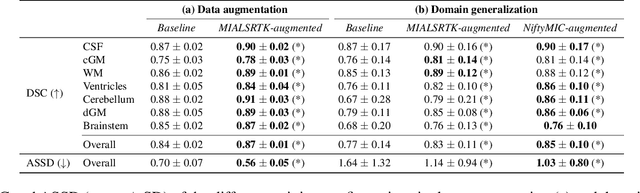
Abstract:Quantitative analysis of in utero human brain development is crucial for abnormal characterization. Magnetic resonance image (MRI) segmentation is therefore an asset for quantitative analysis. However, the development of automated segmentation methods is hampered by the scarce availability of fetal brain MRI annotated datasets and the limited variability within these cohorts. In this context, we propose to leverage the power of fetal brain MRI super-resolution (SR) reconstruction methods to generate multiple reconstructions of a single subject with different parameters, thus as an efficient tuning-free data augmentation strategy. Overall, the latter significantly improves the generalization of segmentation methods over SR pipelines.
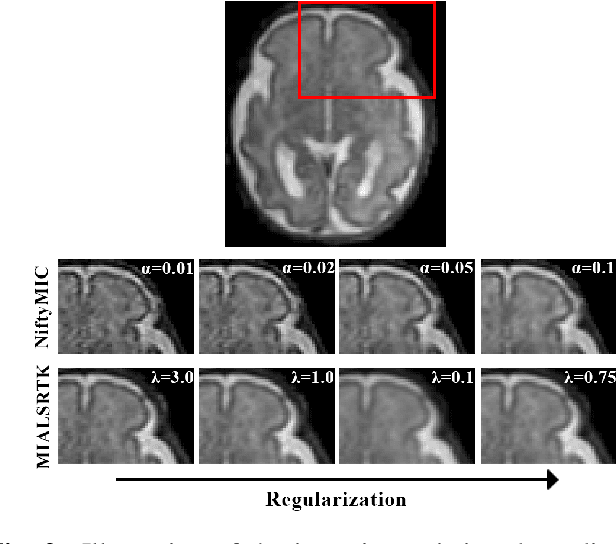
Simulation-based parameter optimization for fetal brain MRI super-resolution reconstruction
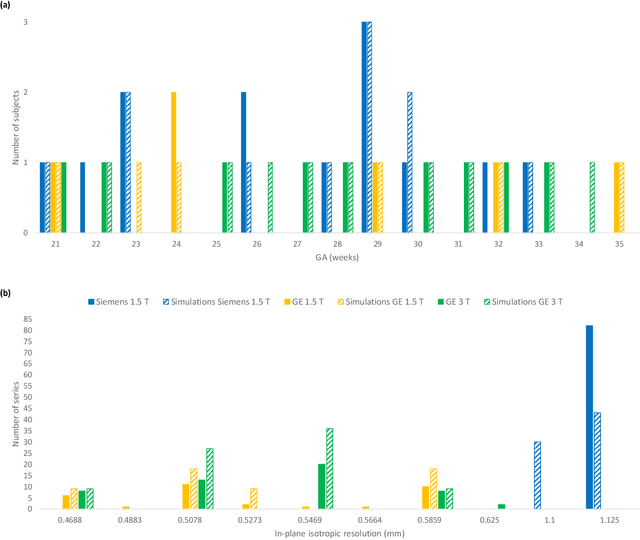
Nov 25, 2022Abstract:In utero fetal brain magnetic resonance images are inherently limited in spatial resolution due to stochastic motion of the fetus. Super-resolution reconstruction methods have become the go-to approach to compute an isotropic motion-free volume of the fetal brain from low-resolution series of 2D thick slices. Such pipelines often rely on an optimization problem with a data fidelity and a regularization term, balanced by a hyperparameter $\alpha$. The lack of ground truth images makes it difficult to adapt $\alpha$ to a given setting of interest in a quantitative manner. In this work, we propose a simulation-based approach to tune $\alpha$ for a given acquisition setting. We focus on two key aspects: the magnetic field strength (1.5T and 3T) and number of LR series used for reconstruction. Our results show that the optimal $\alpha$ significantly improves the performance compared to the default value, across two commonly used SR pipelines. Qualitative validation on clinical data confirms the importance of tuning this parameter to the setting of interest.
Multi-dimensional topological loss for cortical plate segmentation in fetal brain MRI
Aug 16, 2022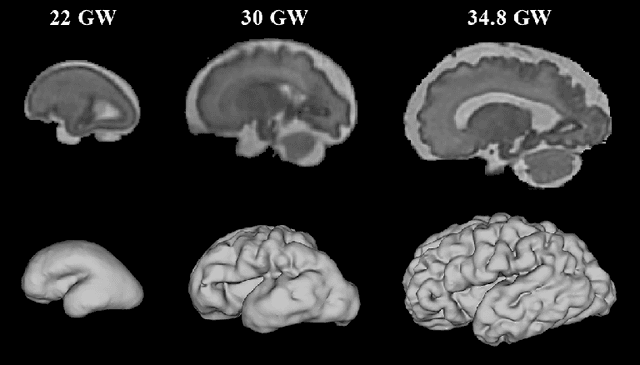
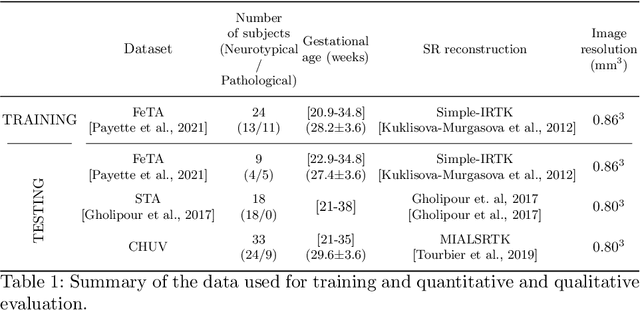
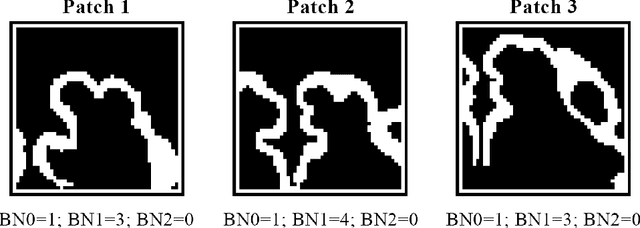
Abstract:The fetal cortical plate (CP) undergoes drastic morphological changes during the in utero development. Therefore, CP growth and folding patterns are key indicator in the assessment of the brain development and maturation. Magnetic resonance imaging (MRI) offers specific insights for the analysis of quantitative imaging biomarkers. Nonetheless, accurate and, more importantly, topologically correct MR image segmentation remains the key baseline to such analysis. In this study, we propose a deep learning segmentation framework for automatic and morphologically consistent segmentation of the CP in fetal brain MRI. Our contribution is two fold. First, we generalized a multi-dimensional topological loss function in order to enhance the topological accuracy. Second, we introduced hole ratio, a new topology-based validation measure that quantifies the size of the topological defects taking into account the size of the structure of interest. Using two publicly available datasets, we quantitatively evaluated our proposed method based on three complementary metrics which are overlap-, distance- and topology-based on 27 fetal brains. Our results evidence that our topology-integrative framework outperforms state-of-the-art training loss functions on super-resolution reconstructed clinical MRI, not only in shape correctness but also in the classical evaluation metrics. Furthermore, results on additional 31 out-of-domain SR reconstructions from clinical acquisitions were qualitatively assessed by three experts. The experts' consensus ranked our TopoCP method as the best segmentation in 100\% of the cases with a high inter-expert agreement. Overall, both quantitative and qualitative results, on a wide range of gestational ages and number of cases, support the generalizability and added value of our topology-guided framework for fetal CP segmentation.
Fetal Brain Tissue Annotation and Segmentation Challenge Results
Apr 20, 2022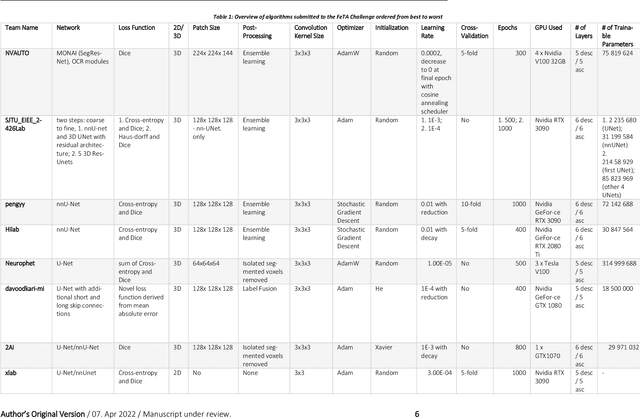
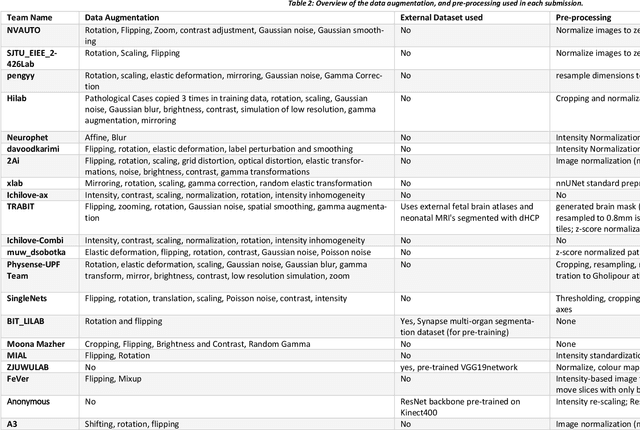
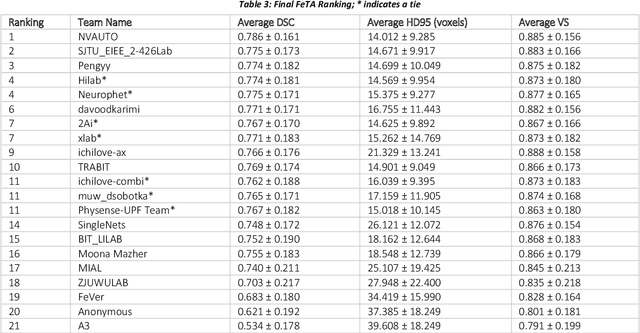
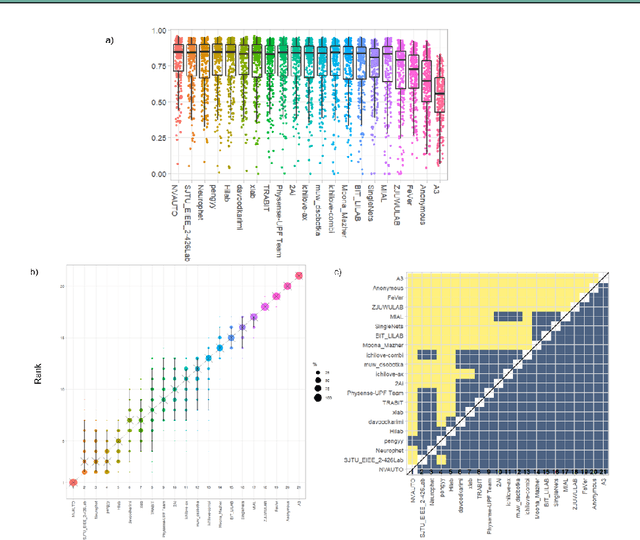
Abstract:In-utero fetal MRI is emerging as an important tool in the diagnosis and analysis of the developing human brain. Automatic segmentation of the developing fetal brain is a vital step in the quantitative analysis of prenatal neurodevelopment both in the research and clinical context. However, manual segmentation of cerebral structures is time-consuming and prone to error and inter-observer variability. Therefore, we organized the Fetal Tissue Annotation (FeTA) Challenge in 2021 in order to encourage the development of automatic segmentation algorithms on an international level. The challenge utilized FeTA Dataset, an open dataset of fetal brain MRI reconstructions segmented into seven different tissues (external cerebrospinal fluid, grey matter, white matter, ventricles, cerebellum, brainstem, deep grey matter). 20 international teams participated in this challenge, submitting a total of 21 algorithms for evaluation. In this paper, we provide a detailed analysis of the results from both a technical and clinical perspective. All participants relied on deep learning methods, mainly U-Nets, with some variability present in the network architecture, optimization, and image pre- and post-processing. The majority of teams used existing medical imaging deep learning frameworks. The main differences between the submissions were the fine tuning done during training, and the specific pre- and post-processing steps performed. The challenge results showed that almost all submissions performed similarly. Four of the top five teams used ensemble learning methods. However, one team's algorithm performed significantly superior to the other submissions, and consisted of an asymmetrical U-Net network architecture. This paper provides a first of its kind benchmark for future automatic multi-tissue segmentation algorithms for the developing human brain in utero.
4D iterative reconstruction of brain fMRI in the moving fetus
Nov 22, 2021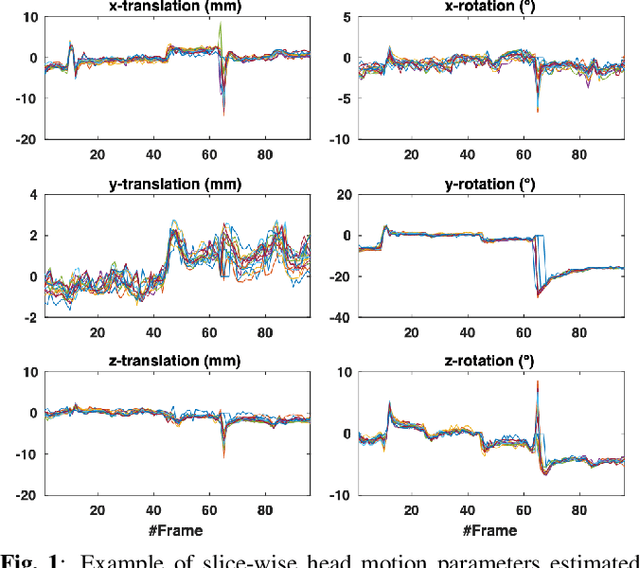
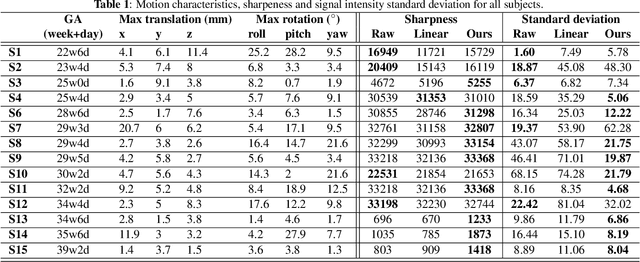
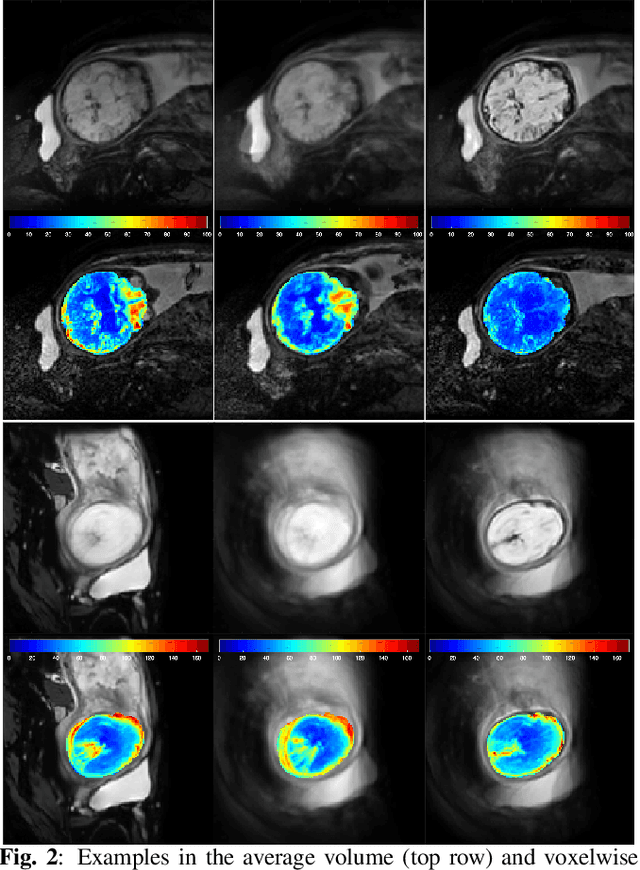
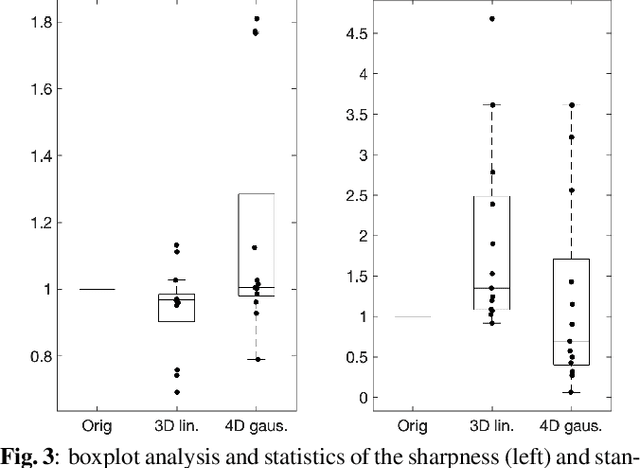
Abstract:Resting-state functional Magnetic Resonance Imaging (fMRI) is a powerful imaging technique for studying functional development of the brain in utero. However, unpredictable and excessive movement of fetuses has limited clinical application since it causes substantial signal fluctuations which can systematically alter observed patterns of functional connectivity. Previous studies have focused on the accurate estimation of the motion parameters in case of large fetal head movement and used a 3D single step interpolation approach at each timepoint to recover motion-free fMRI images. This does not guarantee that the reconstructed image corresponds to the minimum error representation of fMRI time series given the acquired data. Here, we propose a novel technique based on four dimensional iterative reconstruction of the scattered slices acquired during fetal fMRI. The accuracy of the proposed method was quantitatively evaluated on a group of real clinical fMRI fetuses. The results indicate improvements of reconstruction quality compared to the conventional 3D interpolation approach.
Synthetic magnetic resonance images for domain adaptation: Application to fetal brain tissue segmentation
Nov 08, 2021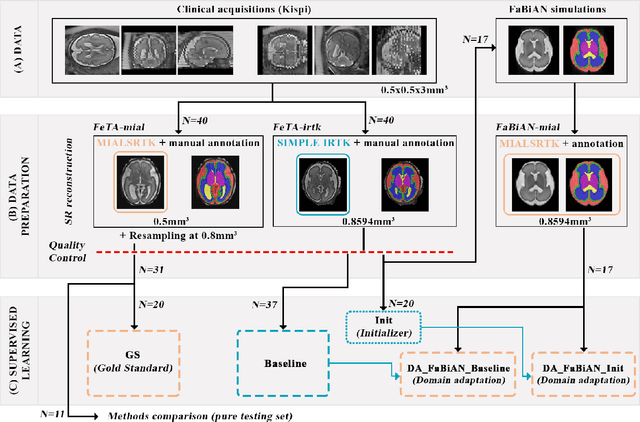
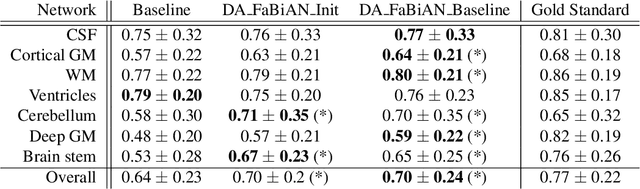
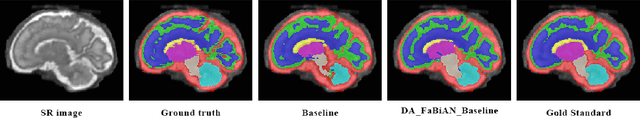
Abstract:The quantitative assessment of the developing human brain in utero is crucial to fully understand neurodevelopment. Thus, automated multi-tissue fetal brain segmentation algorithms are being developed, which in turn require annotated data to be trained. However, the available annotated fetal brain datasets are limited in number and heterogeneity, hampering domain adaptation strategies for robust segmentation. In this context, we use FaBiAN, a Fetal Brain magnetic resonance Acquisition Numerical phantom, to simulate various realistic magnetic resonance images of the fetal brain along with its class labels. We demonstrate that these multiple synthetic annotated data, generated at no cost and further reconstructed using the target super-resolution technique, can be successfully used for domain adaptation of a deep learning method that segments seven brain tissues. Overall, the accuracy of the segmentation is significantly enhanced, especially in the cortical gray matter, the white matter, the cerebellum, the deep gray matter and the brain stem.
FaBiAN: A Fetal Brain magnetic resonance Acquisition Numerical phantom
Sep 06, 2021
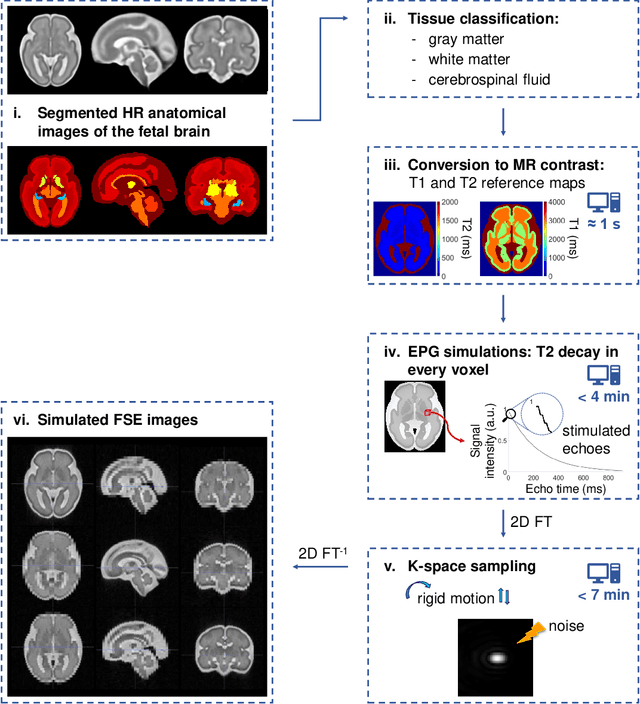

Abstract:Accurate characterization of in utero human brain maturation is critical as it involves complex and interconnected structural and functional processes that may influence health later in life. Magnetic resonance imaging is a powerful tool to investigate equivocal neurological patterns during fetal development. However, the number of acquisitions of satisfactory quality available in this cohort of sensitive subjects remains scarce, thus hindering the validation of advanced image processing techniques. Numerical phantoms can mitigate these limitations by providing a controlled environment with a known ground truth. In this work, we present FaBiAN, an open-source Fetal Brain magnetic resonance Acquisition Numerical phantom that simulates clinical T2-weighted fast spin echo sequences of the fetal brain. This unique tool is based on a general, flexible and realistic setup that includes stochastic fetal movements, thus providing images of the fetal brain throughout maturation comparable to clinical acquisitions. We demonstrate its value to evaluate the robustness and optimize the accuracy of an algorithm for super-resolution fetal brain magnetic resonance imaging from simulated motion-corrupted 2D low-resolution series as compared to a synthetic high-resolution reference volume. We also show that the images generated can complement clinical datasets to support data-intensive deep learning methods for fetal brain tissue segmentation.
A comparison of automatic multi-tissue segmentation methods of the human fetal brain using the FeTA Dataset
Oct 29, 2020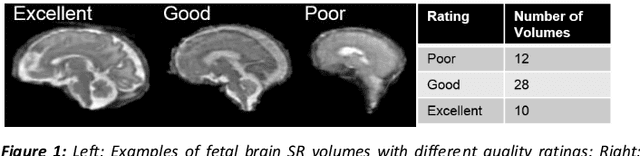
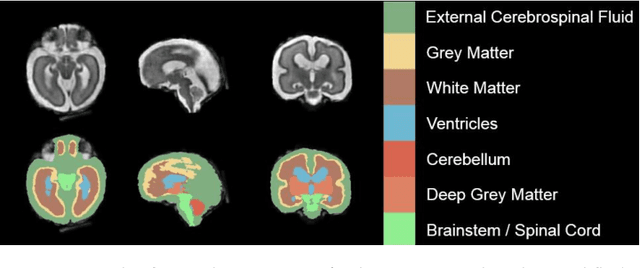
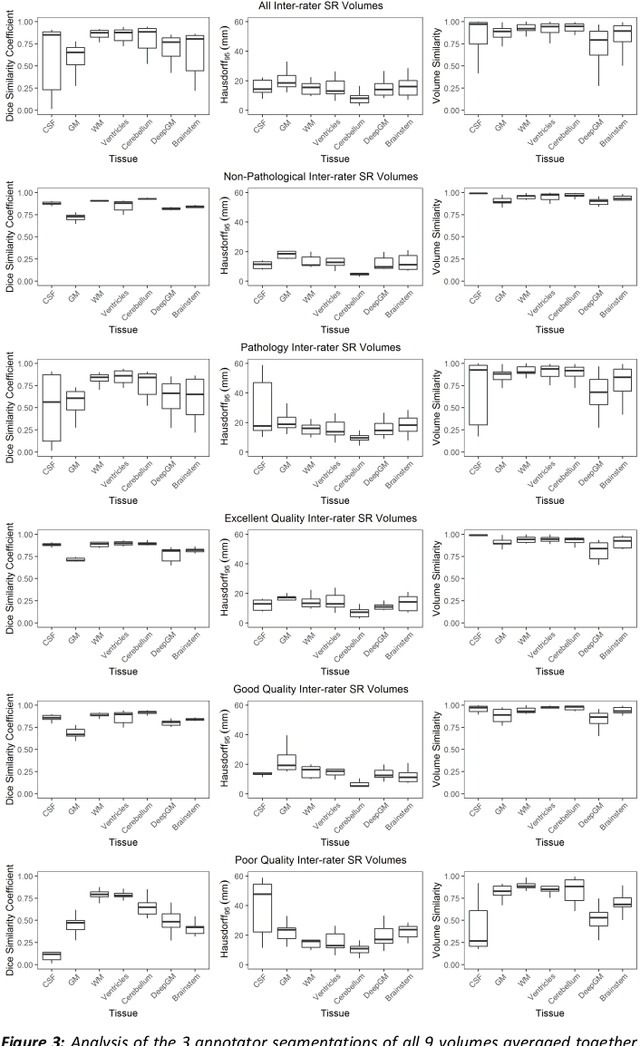
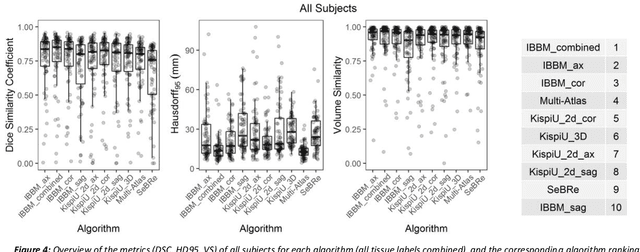
Abstract:It is critical to quantitatively analyse the developing human fetal brain in order to fully understand neurodevelopment in both normal fetuses and those with congenital disorders. To facilitate this analysis, automatic multi-tissue fetal brain segmentation algorithms are needed, which in turn requires open databases of segmented fetal brains. Here we introduce a publicly available database of 50 manually segmented pathological and non-pathological fetal magnetic resonance brain volume reconstructions across a range of gestational ages (20 to 33 weeks) into 7 different tissue categories (external cerebrospinal fluid, grey matter, white matter, ventricles, cerebellum, deep grey matter, brainstem/spinal cord). In addition, we quantitatively evaluate the accuracy of several automatic multi-tissue segmentation algorithms of the developing human fetal brain. Four research groups participated, submitting a total of 10 algorithms, demonstrating the benefits the database for the development of automatic algorithms.
Segmentation of the cortical plate in fetal brain MRI with a topological loss
Oct 23, 2020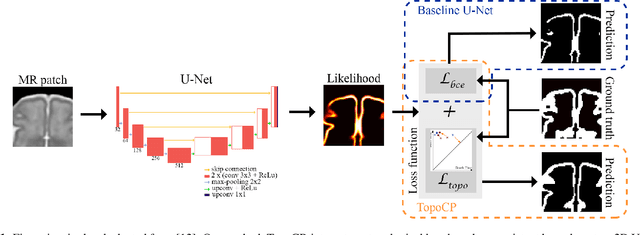

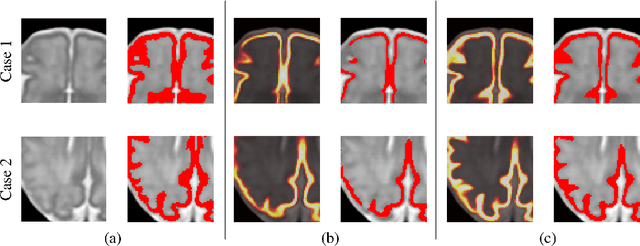

Abstract:The fetal cortical plate undergoes drastic morphological changes throughout early in utero development that can be observed using magnetic resonance (MR) imaging. An accurate MR image segmentation, and more importantly a topologically correct delineation of the cortical gray matter, is a key baseline to perform further quantitative analysis of brain development. In this paper, we propose for the first time the integration of a topological constraint, as an additional loss function, to enhance the morphological consistency of a deep learning-based segmentation of the fetal cortical plate. We quantitatively evaluate our method on 18 fetal brain atlases ranging from 21 to 38 weeks of gestation, showing the significant benefits of our method through all gestational ages as compared to a baseline method. Furthermore, qualitative evaluation by three different experts on 130 randomly selected slices from 26 clinical MRIs evidences the out-performance of our method independently of the MR reconstruction quality.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge