Liu Li
Topology Optimization in Medical Image Segmentation with Fast Euler Characteristic
Jul 31, 2025Abstract:Deep learning-based medical image segmentation techniques have shown promising results when evaluated based on conventional metrics such as the Dice score or Intersection-over-Union. However, these fully automatic methods often fail to meet clinically acceptable accuracy, especially when topological constraints should be observed, e.g., continuous boundaries or closed surfaces. In medical image segmentation, the correctness of a segmentation in terms of the required topological genus sometimes is even more important than the pixel-wise accuracy. Existing topology-aware approaches commonly estimate and constrain the topological structure via the concept of persistent homology (PH). However, these methods are difficult to implement for high dimensional data due to their polynomial computational complexity. To overcome this problem, we propose a novel and fast approach for topology-aware segmentation based on the Euler Characteristic ($\chi$). First, we propose a fast formulation for $\chi$ computation in both 2D and 3D. The scalar $\chi$ error between the prediction and ground-truth serves as the topological evaluation metric. Then we estimate the spatial topology correctness of any segmentation network via a so-called topological violation map, i.e., a detailed map that highlights regions with $\chi$ errors. Finally, the segmentation results from the arbitrary network are refined based on the topological violation maps by a topology-aware correction network. Our experiments are conducted on both 2D and 3D datasets and show that our method can significantly improve topological correctness while preserving pixel-wise segmentation accuracy.
Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge
May 05, 2025



Abstract:Accurate fetal brain tissue segmentation and biometric analysis are essential for studying brain development in utero. The FeTA Challenge 2024 advanced automated fetal brain MRI analysis by introducing biometry prediction as a new task alongside tissue segmentation. For the first time, our diverse multi-centric test set included data from a new low-field (0.55T) MRI dataset. Evaluation metrics were also expanded to include the topology-specific Euler characteristic difference (ED). Sixteen teams submitted segmentation methods, most of which performed consistently across both high- and low-field scans. However, longitudinal trends indicate that segmentation accuracy may be reaching a plateau, with results now approaching inter-rater variability. The ED metric uncovered topological differences that were missed by conventional metrics, while the low-field dataset achieved the highest segmentation scores, highlighting the potential of affordable imaging systems when paired with high-quality reconstruction. Seven teams participated in the biometry task, but most methods failed to outperform a simple baseline that predicted measurements based solely on gestational age, underscoring the challenge of extracting reliable biometric estimates from image data alone. Domain shift analysis identified image quality as the most significant factor affecting model generalization, with super-resolution pipelines also playing a substantial role. Other factors, such as gestational age, pathology, and acquisition site, had smaller, though still measurable, effects. Overall, FeTA 2024 offers a comprehensive benchmark for multi-class segmentation and biometry estimation in fetal brain MRI, underscoring the need for data-centric approaches, improved topological evaluation, and greater dataset diversity to enable clinically robust and generalizable AI tools.
In-Context Learning Distillation for Efficient Few-Shot Fine-Tuning
Dec 17, 2024



Abstract:We applied few-shot in-context learning on the OPT-1.3B model for the natural language inference task and employed knowledge distillation to internalize the context information, reducing model parameter from 1.3B to 125M and achieving a size reduction from 2.5GB to 0.25GB. Compared to using in-context learning alone on similarly sized models, this context distillation approach achieved a nearly 50% improvement in out-of-domain accuracy, demonstrating superior knowledge transfer capabilities over prompt-based methods. Furthermore, this approach reduced memory consumption by up to 60% while delivering a 20% improvement in out-of-domain accuracy compared to conventional pattern-based fine-tuning.
Metasurface-generated large and arbitrary analog convolution kernels for accelerated machine vision
Sep 27, 2024



Abstract:In the rapidly evolving field of artificial intelligence, convolutional neural networks are essential for tackling complex challenges such as machine vision and medical diagnosis. Recently, to address the challenges in processing speed and power consumption of conventional digital convolution operations, many optical components have been suggested to replace the digital convolution layer in the neural network, accelerating various machine vision tasks. Nonetheless, the analog nature of the optical convolution kernel has not been fully explored. Here, we develop a spatial frequency domain training method to create arbitrarily shaped analog convolution kernels using an optical metasurface as the convolution layer, with its receptive field largely surpassing digital convolution kernels. By employing spatial multiplexing, the multiple parallel convolution kernels with both positive and negative weights are generated under the incoherent illumination condition. We experimentally demonstrate a 98.59% classification accuracy on the MNIST dataset, with simulations showing 92.63% and 68.67% accuracy on the Fashion-MNIST and CIFAR-10 datasets with additional digital layers. This work underscores the unique advantage of analog optical convolution, offering a promising avenue to accelerate machine vision tasks, especially in edge devices.
Universal Topology Refinement for Medical Image Segmentation with Polynomial Feature Synthesis
Sep 15, 2024Abstract:Although existing medical image segmentation methods provide impressive pixel-wise accuracy, they often neglect topological correctness, making their segmentations unusable for many downstream tasks. One option is to retrain such models whilst including a topology-driven loss component. However, this is computationally expensive and often impractical. A better solution would be to have a versatile plug-and-play topology refinement method that is compatible with any domain-specific segmentation pipeline. Directly training a post-processing model to mitigate topological errors often fails as such models tend to be biased towards the topological errors of a target segmentation network. The diversity of these errors is confined to the information provided by a labelled training set, which is especially problematic for small datasets. Our method solves this problem by training a model-agnostic topology refinement network with synthetic segmentations that cover a wide variety of topological errors. Inspired by the Stone-Weierstrass theorem, we synthesize topology-perturbation masks with randomly sampled coefficients of orthogonal polynomial bases, which ensures a complete and unbiased representation. Practically, we verified the efficiency and effectiveness of our methods as being compatible with multiple families of polynomial bases, and show evidence that our universal plug-and-play topology refinement network outperforms both existing topology-driven learning-based and post-processing methods. We also show that combining our method with learning-based models provides an effortless add-on, which can further improve the performance of existing approaches.
Stability and Generalizability in SDE Diffusion Models with Measure-Preserving Dynamics
Jun 19, 2024Abstract:Inverse problems describe the process of estimating the causal factors from a set of measurements or data. Mapping of often incomplete or degraded data to parameters is ill-posed, thus data-driven iterative solutions are required, for example when reconstructing clean images from poor signals. Diffusion models have shown promise as potent generative tools for solving inverse problems due to their superior reconstruction quality and their compatibility with iterative solvers. However, most existing approaches are limited to linear inverse problems represented as Stochastic Differential Equations (SDEs). This simplification falls short of addressing the challenging nature of real-world problems, leading to amplified cumulative errors and biases. We provide an explanation for this gap through the lens of measure-preserving dynamics of Random Dynamical Systems (RDS) with which we analyse Temporal Distribution Discrepancy and thus introduce a theoretical framework based on RDS for SDE diffusion models. We uncover several strategies that inherently enhance the stability and generalizability of diffusion models for inverse problems and introduce a novel score-based diffusion framework, the \textbf{D}ynamics-aware S\textbf{D}E \textbf{D}iffusion \textbf{G}enerative \textbf{M}odel (D$^3$GM). The \textit{Measure-preserving property} can return the degraded measurement to the original state despite complex degradation with the RDS concept of \textit{stability}. Our extensive experimental results corroborate the effectiveness of D$^3$GM across multiple benchmarks including a prominent application for inverse problems, magnetic resonance imaging. Code and data will be publicly available.
Weakly Supervised Learning of Cortical Surface Reconstruction from Segmentations
Jun 18, 2024Abstract:Existing learning-based cortical surface reconstruction approaches heavily rely on the supervision of pseudo ground truth (pGT) cortical surfaces for training. Such pGT surfaces are generated by traditional neuroimage processing pipelines, which are time consuming and difficult to generalize well to low-resolution brain MRI, e.g., from fetuses and neonates. In this work, we present CoSeg, a learning-based cortical surface reconstruction framework weakly supervised by brain segmentations without the need for pGT surfaces. CoSeg introduces temporal attention networks to learn time-varying velocity fields from brain MRI for diffeomorphic surface deformations, which fit an initial surface to target cortical surfaces within only 0.11 seconds for each brain hemisphere. A weakly supervised loss is designed to reconstruct pial surfaces by inflating the white surface along the normal direction towards the boundary of the cortical gray matter segmentation. This alleviates partial volume effects and encourages the pial surface to deform into deep and challenging cortical sulci. We evaluate CoSeg on 1,113 adult brain MRI at 1mm and 2mm resolution. CoSeg achieves superior geometric and morphological accuracy compared to existing learning-based approaches. We also verify that CoSeg can extract high-quality cortical surfaces from fetal brain MRI on which traditional pipelines fail to produce acceptable results.
The Developing Human Connectome Project: A Fast Deep Learning-based Pipeline for Neonatal Cortical Surface Reconstruction
May 14, 2024Abstract:The Developing Human Connectome Project (dHCP) aims to explore developmental patterns of the human brain during the perinatal period. An automated processing pipeline has been developed to extract high-quality cortical surfaces from structural brain magnetic resonance (MR) images for the dHCP neonatal dataset. However, the current implementation of the pipeline requires more than 6.5 hours to process a single MRI scan, making it expensive for large-scale neuroimaging studies. In this paper, we propose a fast deep learning (DL) based pipeline for dHCP neonatal cortical surface reconstruction, incorporating DL-based brain extraction, cortical surface reconstruction and spherical projection, as well as GPU-accelerated cortical surface inflation and cortical feature estimation. We introduce a multiscale deformation network to learn diffeomorphic cortical surface reconstruction end-to-end from T2-weighted brain MRI. A fast unsupervised spherical mapping approach is integrated to minimize metric distortions between cortical surfaces and projected spheres. The entire workflow of our DL-based dHCP pipeline completes within only 24 seconds on a modern GPU, which is nearly 1000 times faster than the original dHCP pipeline. Manual quality control demonstrates that for 82.5% of the test samples, our DL-based pipeline produces superior (54.2%) or equal quality (28.3%) cortical surfaces compared to the original dHCP pipeline.
DiffDet4SAR: Diffusion-based Aircraft Target Detection Network for SAR Images
Apr 04, 2024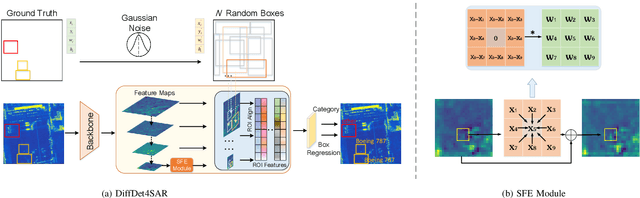
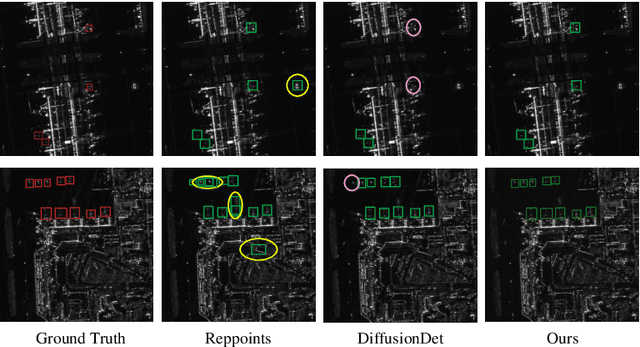
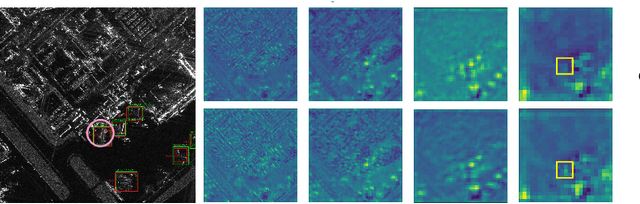
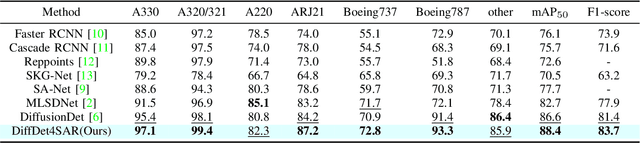
Abstract:Aircraft target detection in SAR images is a challenging task due to the discrete scattering points and severe background clutter interference. Currently, methods with convolution-based or transformer-based paradigms cannot adequately address these issues. In this letter, we explore diffusion models for SAR image aircraft target detection for the first time and propose a novel \underline{Diff}usion-based aircraft target \underline{Det}ection network \underline{for} \underline{SAR} images (DiffDet4SAR). Specifically, the proposed DiffDet4SAR yields two main advantages for SAR aircraft target detection: 1) DiffDet4SAR maps the SAR aircraft target detection task to a denoising diffusion process of bounding boxes without heuristic anchor size selection, effectively enabling large variations in aircraft sizes to be accommodated; and 2) the dedicatedly designed Scattering Feature Enhancement (SFE) module further reduces the clutter intensity and enhances the target saliency during inference. Extensive experimental results on the SAR-AIRcraft-1.0 dataset show that the proposed DiffDet4SAR achieves 88.4\% mAP$_{50}$, outperforming the state-of-the-art methods by 6\%. Code is availabel at \href{https://github.com/JoyeZLearning/DiffDet4SAR}.
Multi-Center Fetal Brain Tissue Annotation (FeTA) Challenge 2022 Results
Feb 08, 2024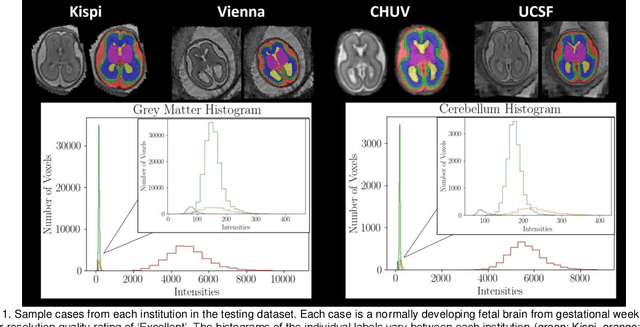
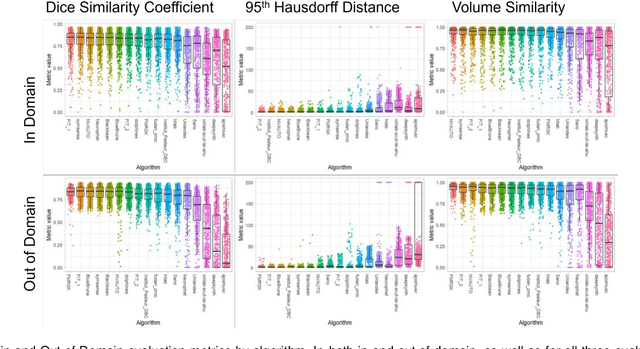
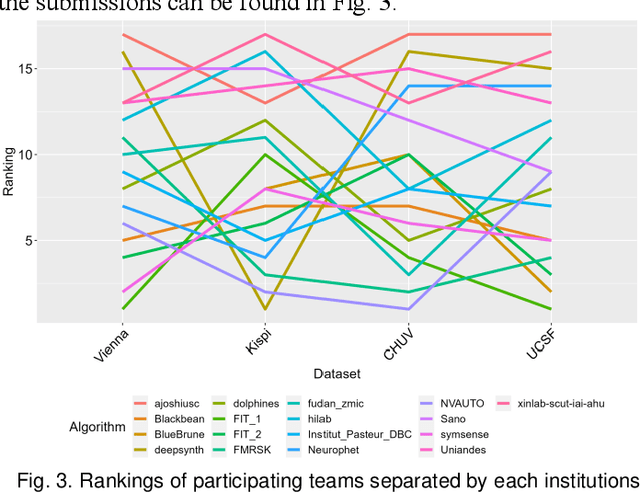
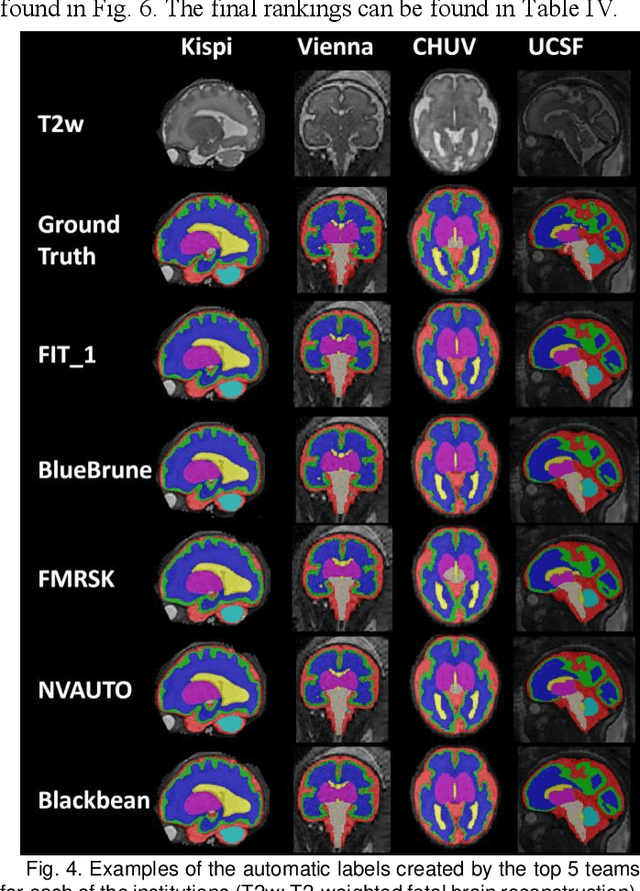
Abstract:Segmentation is a critical step in analyzing the developing human fetal brain. There have been vast improvements in automatic segmentation methods in the past several years, and the Fetal Brain Tissue Annotation (FeTA) Challenge 2021 helped to establish an excellent standard of fetal brain segmentation. However, FeTA 2021 was a single center study, and the generalizability of algorithms across different imaging centers remains unsolved, limiting real-world clinical applicability. The multi-center FeTA Challenge 2022 focuses on advancing the generalizability of fetal brain segmentation algorithms for magnetic resonance imaging (MRI). In FeTA 2022, the training dataset contained images and corresponding manually annotated multi-class labels from two imaging centers, and the testing data contained images from these two imaging centers as well as two additional unseen centers. The data from different centers varied in many aspects, including scanners used, imaging parameters, and fetal brain super-resolution algorithms applied. 16 teams participated in the challenge, and 17 algorithms were evaluated. Here, a detailed overview and analysis of the challenge results are provided, focusing on the generalizability of the submissions. Both in- and out of domain, the white matter and ventricles were segmented with the highest accuracy, while the most challenging structure remains the cerebral cortex due to anatomical complexity. The FeTA Challenge 2022 was able to successfully evaluate and advance generalizability of multi-class fetal brain tissue segmentation algorithms for MRI and it continues to benchmark new algorithms. The resulting new methods contribute to improving the analysis of brain development in utero.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge