Qiang Ma
SwiftVLM: Efficient Vision-Language Model Inference via Cross-Layer Token Bypass
Feb 03, 2026Abstract:Visual token pruning is a promising approach for reducing the computational cost of vision-language models (VLMs), and existing methods often rely on early pruning decisions to improve efficiency. While effective on coarse-grained reasoning tasks, they suffer from significant performance degradation on tasks requiring fine-grained visual details. Through layer-wise analysis, we reveal substantial discrepancies in visual token importance across layers, showing that tokens deemed unimportant at shallow layers can later become highly relevant for text-conditioned reasoning. To avoid irreversible critical information loss caused by premature pruning, we introduce a new pruning paradigm, termed bypass, which preserves unselected visual tokens and forwards them to subsequent pruning stages for re-evaluation. Building on this paradigm, we propose SwiftVLM, a simple and training-free method that performs pruning at model-specific layers with strong visual token selection capability, while enabling independent pruning decisions across layers. Experiments across multiple VLMs and benchmarks demonstrate that SwiftVLM consistently outperforms existing pruning strategies, achieving superior accuracy-efficiency trade-offs and more faithful visual token selection behavior.
MDAFNet: Multiscale Differential Edge and Adaptive Frequency Guided Network for Infrared Small Target Detection
Jan 23, 2026Abstract:Infrared small target detection (IRSTD) plays a crucial role in numerous military and civilian applications. However, existing methods often face the gradual degradation of target edge pixels as the number of network layers increases, and traditional convolution struggles to differentiate between frequency components during feature extraction, leading to low-frequency backgrounds interfering with high-frequency targets and high-frequency noise triggering false detections. To address these limitations, we propose MDAFNet (Multi-scale Differential Edge and Adaptive Frequency Guided Network for Infrared Small Target Detection), which integrates the Multi-Scale Differential Edge (MSDE) module and Dual-Domain Adaptive Feature Enhancement (DAFE) module. The MSDE module, through a multi-scale edge extraction and enhancement mechanism, effectively compensates for the cumulative loss of target edge information during downsampling. The DAFE module combines frequency domain processing mechanisms with simulated frequency decomposition and fusion mechanisms in the spatial domain to effectively improve the network's capability to adaptively enhance high-frequency targets and selectively suppress high-frequency noise. Experimental results on multiple datasets demonstrate the superior detection performance of MDAFNet.
DCCS-Det: Directional Context and Cross-Scale-Aware Detector for Infrared Small Target
Jan 23, 2026Abstract:Infrared small target detection (IRSTD) is critical for applications like remote sensing and surveillance, which aims to identify small, low-contrast targets against complex backgrounds. However, existing methods often struggle with inadequate joint modeling of local-global features (harming target-background discrimination) or feature redundancy and semantic dilution (degrading target representation quality). To tackle these issues, we propose DCCS-Det (Directional Context and Cross-Scale Aware Detector for Infrared Small Target), a novel detector that incorporates a Dual-stream Saliency Enhancement (DSE) block and a Latent-aware Semantic Extraction and Aggregation (LaSEA) module. The DSE block integrates localized perception with direction-aware context aggregation to help capture long-range spatial dependencies and local details. On this basis, the LaSEA module mitigates feature degradation via cross-scale feature extraction and random pooling sampling strategies, enhancing discriminative features and suppressing noise. Extensive experiments show that DCCS-Det achieves state-of-the-art detection accuracy with competitive efficiency across multiple datasets. Ablation studies further validate the contributions of DSE and LaSEA in improving target perception and feature representation under complex scenarios. \href{https://huggingface.co/InPeerReview/InfraredSmallTargetDetection-IRSTD.DCCS}{DCCS-Det Official Code is Available Here!}
SpotVLM: Cloud-edge Collaborative Real-time VLM based on Context Transfer
Aug 18, 2025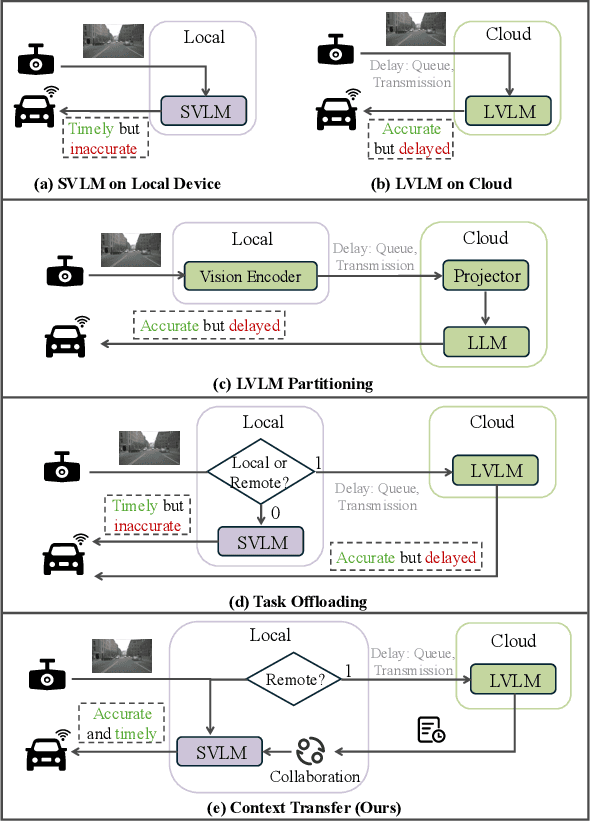
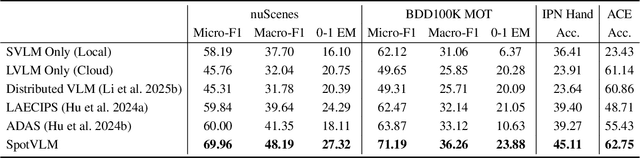
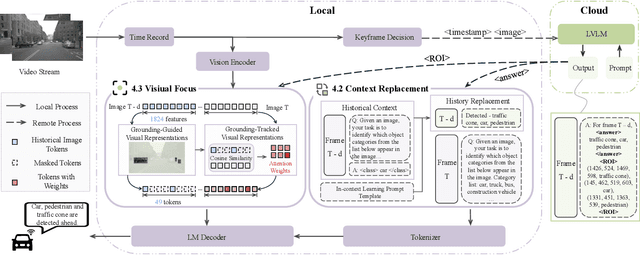
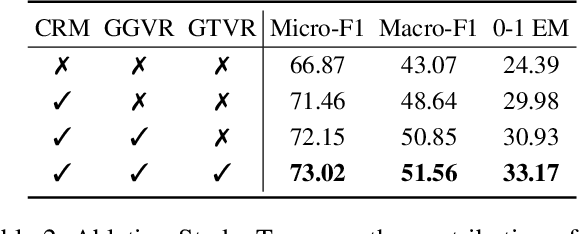
Abstract:Vision-Language Models (VLMs) are increasingly deployed in real-time applications such as autonomous driving and human-computer interaction, which demand fast and reliable responses based on accurate perception. To meet these requirements, existing systems commonly employ cloud-edge collaborative architectures, such as partitioned Large Vision-Language Models (LVLMs) or task offloading strategies between Large and Small Vision-Language Models (SVLMs). However, these methods fail to accommodate cloud latency fluctuations and overlook the full potential of delayed but accurate LVLM responses. In this work, we propose a novel cloud-edge collaborative paradigm for VLMs, termed Context Transfer, which treats the delayed outputs of LVLMs as historical context to provide real-time guidance for SVLMs inference. Based on this paradigm, we design SpotVLM, which incorporates both context replacement and visual focus modules to refine historical textual input and enhance visual grounding consistency. Extensive experiments on three real-time vision tasks across four datasets demonstrate the effectiveness of the proposed framework. The new paradigm lays the groundwork for more effective and latency-aware collaboration strategies in future VLM systems.
OpenMoCap: Rethinking Optical Motion Capture under Real-world Occlusion
Aug 18, 2025Abstract:Optical motion capture is a foundational technology driving advancements in cutting-edge fields such as virtual reality and film production. However, system performance suffers severely under large-scale marker occlusions common in real-world applications. An in-depth analysis identifies two primary limitations of current models: (i) the lack of training datasets accurately reflecting realistic marker occlusion patterns, and (ii) the absence of training strategies designed to capture long-range dependencies among markers. To tackle these challenges, we introduce the CMU-Occlu dataset, which incorporates ray tracing techniques to realistically simulate practical marker occlusion patterns. Furthermore, we propose OpenMoCap, a novel motion-solving model designed specifically for robust motion capture in environments with significant occlusions. Leveraging a marker-joint chain inference mechanism, OpenMoCap enables simultaneous optimization and construction of deep constraints between markers and joints. Extensive comparative experiments demonstrate that OpenMoCap consistently outperforms competing methods across diverse scenarios, while the CMU-Occlu dataset opens the door for future studies in robust motion solving. The proposed OpenMoCap is integrated into the MoSen MoCap system for practical deployment. The code is released at: https://github.com/qianchen214/OpenMoCap.
Topology Optimization in Medical Image Segmentation with Fast Euler Characteristic
Jul 31, 2025Abstract:Deep learning-based medical image segmentation techniques have shown promising results when evaluated based on conventional metrics such as the Dice score or Intersection-over-Union. However, these fully automatic methods often fail to meet clinically acceptable accuracy, especially when topological constraints should be observed, e.g., continuous boundaries or closed surfaces. In medical image segmentation, the correctness of a segmentation in terms of the required topological genus sometimes is even more important than the pixel-wise accuracy. Existing topology-aware approaches commonly estimate and constrain the topological structure via the concept of persistent homology (PH). However, these methods are difficult to implement for high dimensional data due to their polynomial computational complexity. To overcome this problem, we propose a novel and fast approach for topology-aware segmentation based on the Euler Characteristic ($\chi$). First, we propose a fast formulation for $\chi$ computation in both 2D and 3D. The scalar $\chi$ error between the prediction and ground-truth serves as the topological evaluation metric. Then we estimate the spatial topology correctness of any segmentation network via a so-called topological violation map, i.e., a detailed map that highlights regions with $\chi$ errors. Finally, the segmentation results from the arbitrary network are refined based on the topological violation maps by a topology-aware correction network. Our experiments are conducted on both 2D and 3D datasets and show that our method can significantly improve topological correctness while preserving pixel-wise segmentation accuracy.
XLRS-Bench: Could Your Multimodal LLMs Understand Extremely Large Ultra-High-Resolution Remote Sensing Imagery?
Mar 31, 2025Abstract:The astonishing breakthrough of multimodal large language models (MLLMs) has necessitated new benchmarks to quantitatively assess their capabilities, reveal their limitations, and indicate future research directions. However, this is challenging in the context of remote sensing (RS), since the imagery features ultra-high resolution that incorporates extremely complex semantic relationships. Existing benchmarks usually adopt notably smaller image sizes than real-world RS scenarios, suffer from limited annotation quality, and consider insufficient dimensions of evaluation. To address these issues, we present XLRS-Bench: a comprehensive benchmark for evaluating the perception and reasoning capabilities of MLLMs in ultra-high-resolution RS scenarios. XLRS-Bench boasts the largest average image size (8500$\times$8500) observed thus far, with all evaluation samples meticulously annotated manually, assisted by a novel semi-automatic captioner on ultra-high-resolution RS images. On top of the XLRS-Bench, 16 sub-tasks are defined to evaluate MLLMs' 10 kinds of perceptual capabilities and 6 kinds of reasoning capabilities, with a primary emphasis on advanced cognitive processes that facilitate real-world decision-making and the capture of spatiotemporal changes. The results of both general and RS-focused MLLMs on XLRS-Bench indicate that further efforts are needed for real-world RS applications. We have open-sourced XLRS-Bench to support further research in developing more powerful MLLMs for remote sensing.
Ensemble ToT of LLMs and Its Application to Automatic Grading System for Supporting Self-Learning
Feb 23, 2025Abstract:Providing students with detailed and timely grading feedback is essential for self-learning. While existing LLM-based grading systems are promising, most of them rely on one single model, which limits their performance. To address this, we propose Ensemble Tree-of-Thought (ToT), a framework that enhances LLM outputs by integrating multiple models. Using this framework, we develop a grading system. Ensemble ToT follows three steps: (1) analyzing LLM performance, (2) generating candidate answers, and (3) refining them into a final result. Based on this, our grading system first evaluates the grading tendencies of LLMs, then generates multiple results, and finally integrates them via a simulated debate. Experimental results demonstrate our approach's ability to provide accurate and explainable grading by effectively coordinating multiple LLMs.
CAMEF: Causal-Augmented Multi-Modality Event-Driven Financial Forecasting by Integrating Time Series Patterns and Salient Macroeconomic Announcements
Feb 07, 2025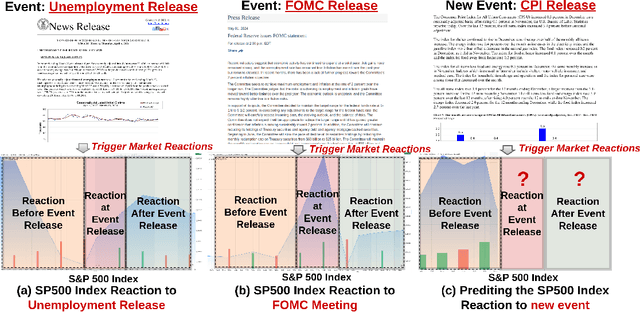

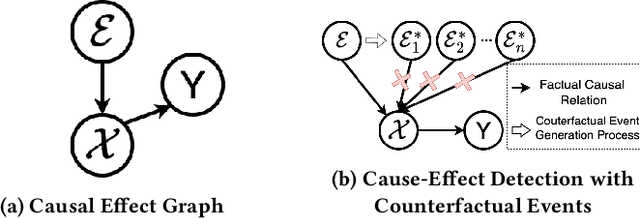

Abstract:Accurately forecasting the impact of macroeconomic events is critical for investors and policymakers. Salient events like monetary policy decisions and employment reports often trigger market movements by shaping expectations of economic growth and risk, thereby establishing causal relationships between events and market behavior. Existing forecasting methods typically focus either on textual analysis or time-series modeling, but fail to capture the multi-modal nature of financial markets and the causal relationship between events and price movements. To address these gaps, we propose CAMEF (Causal-Augmented Multi-Modality Event-Driven Financial Forecasting), a multi-modality framework that effectively integrates textual and time-series data with a causal learning mechanism and an LLM-based counterfactual event augmentation technique for causal-enhanced financial forecasting. Our contributions include: (1) a multi-modal framework that captures causal relationships between policy texts and historical price data; (2) a new financial dataset with six types of macroeconomic releases from 2008 to April 2024, and high-frequency real trading data for five key U.S. financial assets; and (3) an LLM-based counterfactual event augmentation strategy. We compare CAMEF to state-of-the-art transformer-based time-series and multi-modal baselines, and perform ablation studies to validate the effectiveness of the causal learning mechanism and event types.
Universal Topology Refinement for Medical Image Segmentation with Polynomial Feature Synthesis
Sep 15, 2024Abstract:Although existing medical image segmentation methods provide impressive pixel-wise accuracy, they often neglect topological correctness, making their segmentations unusable for many downstream tasks. One option is to retrain such models whilst including a topology-driven loss component. However, this is computationally expensive and often impractical. A better solution would be to have a versatile plug-and-play topology refinement method that is compatible with any domain-specific segmentation pipeline. Directly training a post-processing model to mitigate topological errors often fails as such models tend to be biased towards the topological errors of a target segmentation network. The diversity of these errors is confined to the information provided by a labelled training set, which is especially problematic for small datasets. Our method solves this problem by training a model-agnostic topology refinement network with synthetic segmentations that cover a wide variety of topological errors. Inspired by the Stone-Weierstrass theorem, we synthesize topology-perturbation masks with randomly sampled coefficients of orthogonal polynomial bases, which ensures a complete and unbiased representation. Practically, we verified the efficiency and effectiveness of our methods as being compatible with multiple families of polynomial bases, and show evidence that our universal plug-and-play topology refinement network outperforms both existing topology-driven learning-based and post-processing methods. We also show that combining our method with learning-based models provides an effortless add-on, which can further improve the performance of existing approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge