Thomas Sanchez
Fetpype: An Open-Source Pipeline for Reproducible Fetal Brain MRI Analysis
Dec 19, 2025Abstract:Fetal brain Magnetic Resonance Imaging (MRI) is crucial for assessing neurodevelopment in utero. However, analyzing this data presents significant challenges due to fetal motion, low signal-to-noise ratio, and the need for complex multi-step processing, including motion correction, super-resolution reconstruction, segmentation, and surface extraction. While various specialized tools exist for individual steps, integrating them into robust, reproducible, and user-friendly workflows that go from raw images to processed volumes is not straightforward. This lack of standardization hinders reproducibility across studies and limits the adoption of advanced analysis techniques for researchers and clinicians. To address these challenges, we introduce Fetpype, an open-source Python library designed to streamline and standardize the preprocessing and analysis of T2-weighted fetal brain MRI data. Fetpype is publicly available on GitHub at https://github.com/fetpype/fetpype.
Causal Attribution of Model Performance Gaps in Medical Imaging Under Distribution Shifts
Dec 09, 2025Abstract:Deep learning models for medical image segmentation suffer significant performance drops due to distribution shifts, but the causal mechanisms behind these drops remain poorly understood. We extend causal attribution frameworks to high-dimensional segmentation tasks, quantifying how acquisition protocols and annotation variability independently contribute to performance degradation. We model the data-generating process through a causal graph and employ Shapley values to fairly attribute performance changes to individual mechanisms. Our framework addresses unique challenges in medical imaging: high-dimensional outputs, limited samples, and complex mechanism interactions. Validation on multiple sclerosis (MS) lesion segmentation across 4 centers and 7 annotators reveals context-dependent failure modes: annotation protocol shifts dominate when crossing annotators (7.4% $\pm$ 8.9% DSC attribution), while acquisition shifts dominate when crossing imaging centers (6.5% $\pm$ 9.1%). This mechanism-specific quantification enables practitioners to prioritize targeted interventions based on deployment context.
Meta-learning Slice-to-Volume Reconstruction in Fetal Brain MRI using Implicit Neural Representations
May 14, 2025



Abstract:High-resolution slice-to-volume reconstruction (SVR) from multiple motion-corrupted low-resolution 2D slices constitutes a critical step in image-based diagnostics of moving subjects, such as fetal brain Magnetic Resonance Imaging (MRI). Existing solutions struggle with image artifacts and severe subject motion or require slice pre-alignment to achieve satisfying reconstruction performance. We propose a novel SVR method to enable fast and accurate MRI reconstruction even in cases of severe image and motion corruption. Our approach performs motion correction, outlier handling, and super-resolution reconstruction with all operations being entirely based on implicit neural representations. The model can be initialized with task-specific priors through fully self-supervised meta-learning on either simulated or real-world data. In extensive experiments including over 480 reconstructions of simulated and clinical MRI brain data from different centers, we prove the utility of our method in cases of severe subject motion and image artifacts. Our results demonstrate improvements in reconstruction quality, especially in the presence of severe motion, compared to state-of-the-art methods, and up to 50% reduction in reconstruction time.
Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge
May 05, 2025



Abstract:Accurate fetal brain tissue segmentation and biometric analysis are essential for studying brain development in utero. The FeTA Challenge 2024 advanced automated fetal brain MRI analysis by introducing biometry prediction as a new task alongside tissue segmentation. For the first time, our diverse multi-centric test set included data from a new low-field (0.55T) MRI dataset. Evaluation metrics were also expanded to include the topology-specific Euler characteristic difference (ED). Sixteen teams submitted segmentation methods, most of which performed consistently across both high- and low-field scans. However, longitudinal trends indicate that segmentation accuracy may be reaching a plateau, with results now approaching inter-rater variability. The ED metric uncovered topological differences that were missed by conventional metrics, while the low-field dataset achieved the highest segmentation scores, highlighting the potential of affordable imaging systems when paired with high-quality reconstruction. Seven teams participated in the biometry task, but most methods failed to outperform a simple baseline that predicted measurements based solely on gestational age, underscoring the challenge of extracting reliable biometric estimates from image data alone. Domain shift analysis identified image quality as the most significant factor affecting model generalization, with super-resolution pipelines also playing a substantial role. Other factors, such as gestational age, pathology, and acquisition site, had smaller, though still measurable, effects. Overall, FeTA 2024 offers a comprehensive benchmark for multi-class segmentation and biometry estimation in fetal brain MRI, underscoring the need for data-centric approaches, improved topological evaluation, and greater dataset diversity to enable clinically robust and generalizable AI tools.
Automatic quality control in multi-centric fetal brain MRI super-resolution reconstruction
Mar 13, 2025Abstract:Quality control (QC) has long been considered essential to guarantee the reliability of neuroimaging studies. It is particularly important for fetal brain MRI, where acquisitions and image processing techniques are less standardized than in adult imaging. In this work, we focus on automated quality control of super-resolution reconstruction (SRR) volumes of fetal brain MRI, an important processing step where multiple stacks of thick 2D slices are registered together and combined to build a single, isotropic and artifact-free T2 weighted volume. We propose FetMRQC$_{SR}$, a machine-learning method that extracts more than 100 image quality metrics to predict image quality scores using a random forest model. This approach is well suited to a problem that is high dimensional, with highly heterogeneous data and small datasets. We validate FetMRQC$_{SR}$ in an out-of-domain (OOD) setting and report high performance (ROC AUC = 0.89), even when faced with data from an unknown site or SRR method. We also investigate failure cases and show that they occur in $45\%$ of the images due to ambiguous configurations for which the rating from the expert is arguable. These results are encouraging and illustrate how a non deep learning-based method like FetMRQC$_{SR}$ is well suited to this multifaceted problem. Our tool, along with all the code used to generate, train and evaluate the model will be released upon acceptance of the paper.
Maximizing domain generalization in fetal brain tissue segmentation: the role of synthetic data generation, intensity clustering and real image fine-tuning
Nov 11, 2024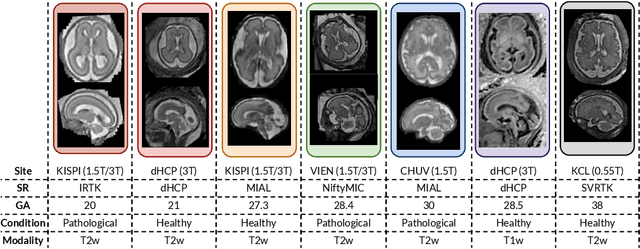
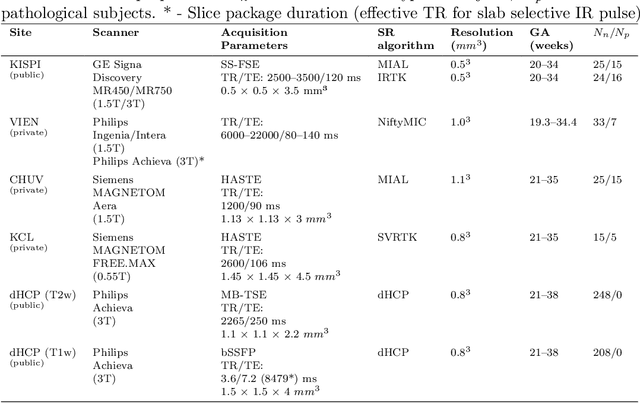
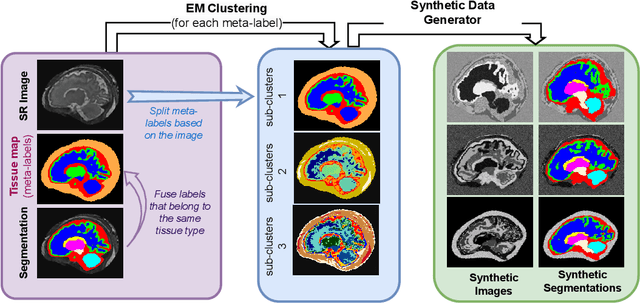
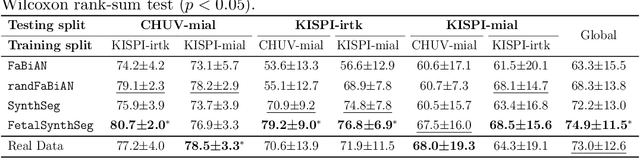
Abstract:Fetal brain tissue segmentation in magnetic resonance imaging (MRI) is a crucial tool that supports the understanding of neurodevelopment, yet it faces challenges due to the heterogeneity of data coming from different scanners and settings, and due to data scarcity. Recent approaches based on domain randomization, like SynthSeg, have shown a great potential for single source domain generalization, by simulating images with randomized contrast and image resolution from the label maps. In this work, we investigate how to maximize the out-of-domain (OOD) generalization potential of SynthSeg-based methods in fetal brain MRI. Specifically, when studying data generation, we demonstrate that the simple Gaussian mixture models used in SynthSeg enable more robust OOD generalization than physics-informed generation methods. We also investigate how intensity clustering can help create more faithful synthetic images, and observe that it is key to achieving a non-trivial OOD generalization capability when few label classes are available. Finally, by combining for the first time SynthSeg with modern fine-tuning approaches based on weight averaging, we show that fine-tuning a model pre-trained on synthetic data on a few real image-segmentation pairs in a new domain can lead to improvements in the target domain, but also in other domains. We summarize our findings as five key recommendations that we believe can guide practitioners who would like to develop SynthSeg-based approaches in other organs or modalities.
Improving cross-domain brain tissue segmentation in fetal MRI with synthetic data
Mar 22, 2024Abstract:Segmentation of fetal brain tissue from magnetic resonance imaging (MRI) plays a crucial role in the study of in utero neurodevelopment. However, automated tools face substantial domain shift challenges as they must be robust to highly heterogeneous clinical data, often limited in numbers and lacking annotations. Indeed, high variability of the fetal brain morphology, MRI acquisition parameters, and superresolution reconstruction (SR) algorithms adversely affect the model's performance when evaluated out-of-domain. In this work, we introduce FetalSynthSeg, a domain randomization method to segment fetal brain MRI, inspired by SynthSeg. Our results show that models trained solely on synthetic data outperform models trained on real data in out-ofdomain settings, validated on a 120-subject cross-domain dataset. Furthermore, we extend our evaluation to 40 subjects acquired using lowfield (0.55T) MRI and reconstructed with novel SR models, showcasing robustness across different magnetic field strengths and SR algorithms. Leveraging a generative synthetic approach, we tackle the domain shift problem in fetal brain MRI and offer compelling prospects for applications in fields with limited and highly heterogeneous data.
Learning to sample in Cartesian MRI
Dec 07, 2023



Abstract:Despite its exceptional soft tissue contrast, Magnetic Resonance Imaging (MRI) faces the challenge of long scanning times compared to other modalities like X-ray radiography. Shortening scanning times is crucial in clinical settings, as it increases patient comfort, decreases examination costs and improves throughput. Recent advances in compressed sensing (CS) and deep learning allow accelerated MRI acquisition by reconstructing high-quality images from undersampled data. While reconstruction algorithms have received most of the focus, designing acquisition trajectories to optimize reconstruction quality remains an open question. This thesis explores two approaches to address this gap in the context of Cartesian MRI. First, we propose two algorithms, lazy LBCS and stochastic LBCS, that significantly improve upon G\"ozc\"u et al.'s greedy learning-based CS (LBCS) approach. These algorithms scale to large, clinically relevant scenarios like multi-coil 3D MR and dynamic MRI, previously inaccessible to LBCS. Additionally, we demonstrate that generative adversarial networks (GANs) can serve as a natural criterion for adaptive sampling by leveraging variance in the measurement domain to guide acquisition. Second, we delve into the underlying structures or assumptions that enable mask design algorithms to perform well in practice. Our experiments reveal that state-of-the-art deep reinforcement learning (RL) approaches, while capable of adaptation and long-horizon planning, offer only marginal improvements over stochastic LBCS, which is neither adaptive nor does long-term planning. Altogether, our findings suggest that stochastic LBCS and similar methods represent promising alternatives to deep RL. They shine in particular by their scalability and computational efficiency and could be key in the deployment of optimized acquisition trajectories in Cartesian MRI.
FetMRQC: an open-source machine learning framework for multi-centric fetal brain MRI quality control
Nov 08, 2023Abstract:Fetal brain MRI is becoming an increasingly relevant complement to neurosonography for perinatal diagnosis, allowing fundamental insights into fetal brain development throughout gestation. However, uncontrolled fetal motion and heterogeneity in acquisition protocols lead to data of variable quality, potentially biasing the outcome of subsequent studies. We present FetMRQC, an open-source machine-learning framework for automated image quality assessment and quality control that is robust to domain shifts induced by the heterogeneity of clinical data. FetMRQC extracts an ensemble of quality metrics from unprocessed anatomical MRI and combines them to predict experts' ratings using random forests. We validate our framework on a pioneeringly large and diverse dataset of more than 1600 manually rated fetal brain T2-weighted images from four clinical centers and 13 different scanners. Our study shows that FetMRQC's predictions generalize well to unseen data while being interpretable. FetMRQC is a step towards more robust fetal brain neuroimaging, which has the potential to shed new insights on the developing human brain.
FetMRQC: Automated Quality Control for fetal brain MRI
Apr 12, 2023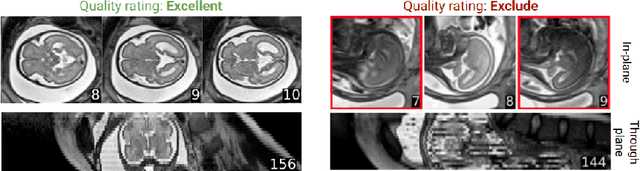
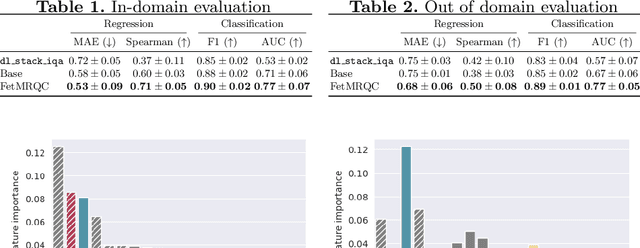
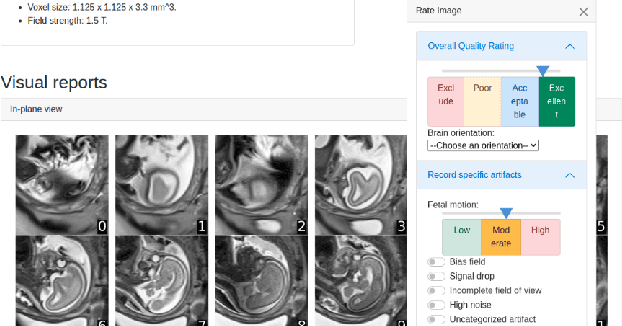
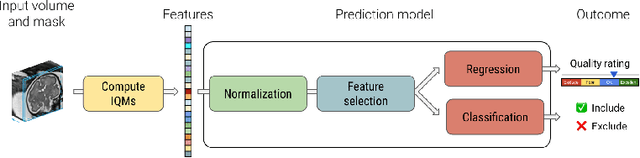
Abstract:Quality control (QC) has long been considered essential to guarantee the reliability of neuroimaging studies. It is particularly important for fetal brain MRI, where large and unpredictable fetal motion can lead to substantial artifacts in the acquired images. Existing methods for fetal brain quality assessment operate at the \textit{slice} level, and fail to get a comprehensive picture of the quality of an image, that can only be achieved by looking at the \textit{entire} brain volume. In this work, we propose FetMRQC, a machine learning framework for automated image quality assessment tailored to fetal brain MRI, which extracts an ensemble of quality metrics that are then used to predict experts' ratings. Based on the manual ratings of more than 1000 low-resolution stacks acquired across two different institutions, we show that, compared with existing quality metrics, FetMRQC is able to generalize out-of-domain, while being interpretable and data efficient. We also release a novel manual quality rating tool designed to facilitate and optimize quality rating of fetal brain images. Our tool, along with all the code to generate, train and evaluate the model will be released upon acceptance of the paper.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge