Oscar Esteban
Department of Radiology, Université de Lausanne
FetMRQC: an open-source machine learning framework for multi-centric fetal brain MRI quality control
Nov 08, 2023Abstract:Fetal brain MRI is becoming an increasingly relevant complement to neurosonography for perinatal diagnosis, allowing fundamental insights into fetal brain development throughout gestation. However, uncontrolled fetal motion and heterogeneity in acquisition protocols lead to data of variable quality, potentially biasing the outcome of subsequent studies. We present FetMRQC, an open-source machine-learning framework for automated image quality assessment and quality control that is robust to domain shifts induced by the heterogeneity of clinical data. FetMRQC extracts an ensemble of quality metrics from unprocessed anatomical MRI and combines them to predict experts' ratings using random forests. We validate our framework on a pioneeringly large and diverse dataset of more than 1600 manually rated fetal brain T2-weighted images from four clinical centers and 13 different scanners. Our study shows that FetMRQC's predictions generalize well to unseen data while being interpretable. FetMRQC is a step towards more robust fetal brain neuroimaging, which has the potential to shed new insights on the developing human brain.
FetMRQC: Automated Quality Control for fetal brain MRI
Apr 12, 2023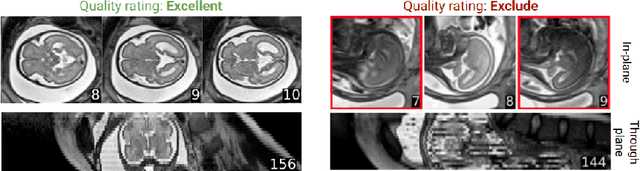
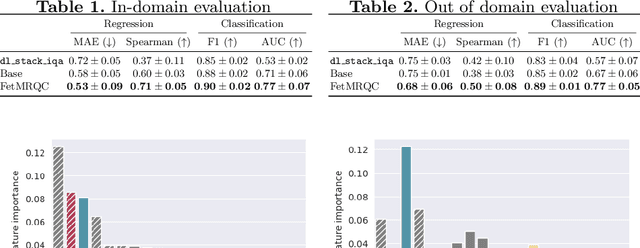
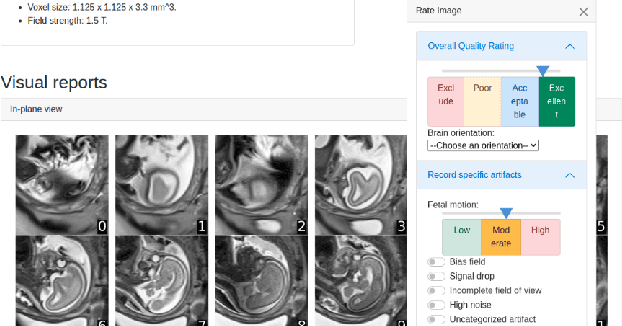
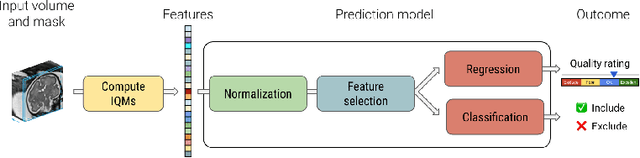
Abstract:Quality control (QC) has long been considered essential to guarantee the reliability of neuroimaging studies. It is particularly important for fetal brain MRI, where large and unpredictable fetal motion can lead to substantial artifacts in the acquired images. Existing methods for fetal brain quality assessment operate at the \textit{slice} level, and fail to get a comprehensive picture of the quality of an image, that can only be achieved by looking at the \textit{entire} brain volume. In this work, we propose FetMRQC, a machine learning framework for automated image quality assessment tailored to fetal brain MRI, which extracts an ensemble of quality metrics that are then used to predict experts' ratings. Based on the manual ratings of more than 1000 low-resolution stacks acquired across two different institutions, we show that, compared with existing quality metrics, FetMRQC is able to generalize out-of-domain, while being interpretable and data efficient. We also release a novel manual quality rating tool designed to facilitate and optimize quality rating of fetal brain images. Our tool, along with all the code to generate, train and evaluate the model will be released upon acceptance of the paper.
Differentiable programming for functional connectomics
May 31, 2022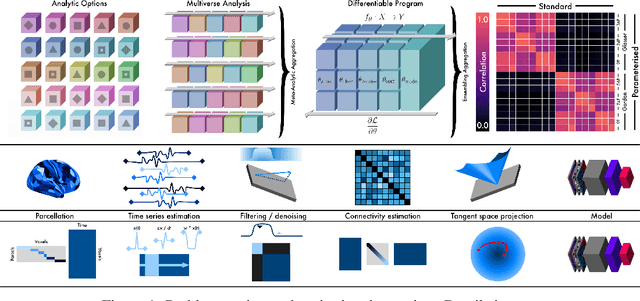
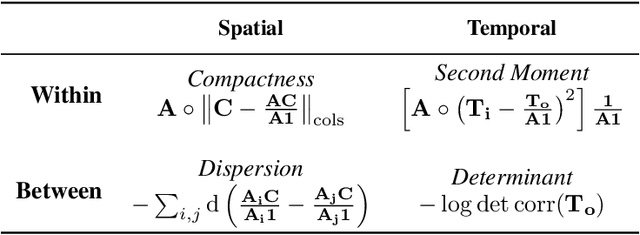
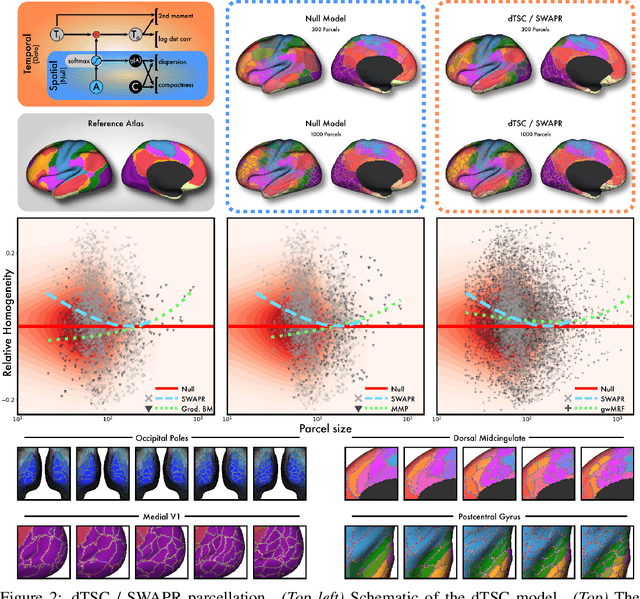
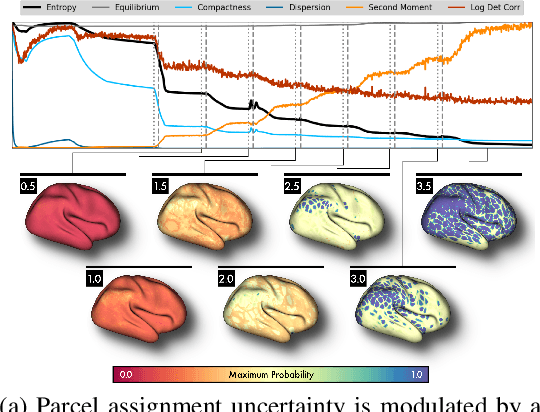
Abstract:Mapping the functional connectome has the potential to uncover key insights into brain organisation. However, existing workflows for functional connectomics are limited in their adaptability to new data, and principled workflow design is a challenging combinatorial problem. We introduce a new analytic paradigm and software toolbox that implements common operations used in functional connectomics as fully differentiable processing blocks. Under this paradigm, workflow configurations exist as reparameterisations of a differentiable functional that interpolates them. The differentiable program that we envision occupies a niche midway between traditional pipelines and end-to-end neural networks, combining the glass-box tractability and domain knowledge of the former with the amenability to optimisation of the latter. In this preliminary work, we provide a proof of concept for differentiable connectomics, demonstrating the capacity of our processing blocks both to recapitulate canonical knowledge in neuroscience and to make new discoveries in an unsupervised setting. Our differentiable modules are competitive with state-of-the-art methods in problem domains including functional parcellation, denoising, and covariance modelling. Taken together, our results and software demonstrate the promise of differentiable programming for functional connectomics.
Weak labels and anatomical knowledge: making deep learning practical for intracranial aneurysm detection in TOF-MRA
Mar 10, 2021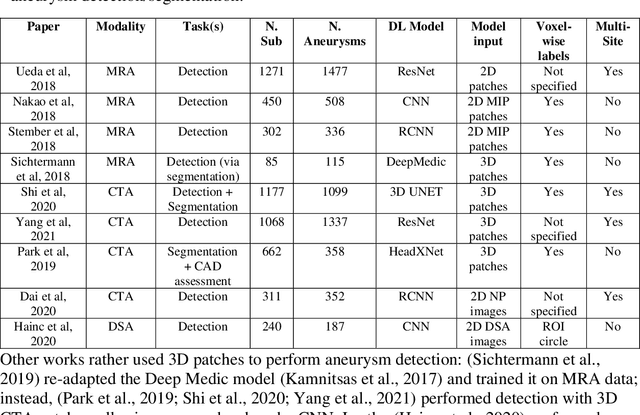
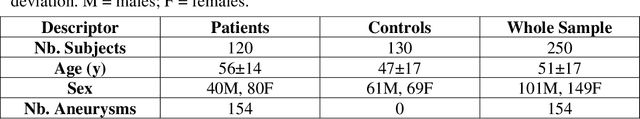
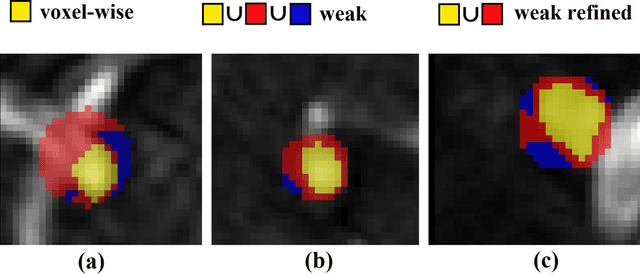
Abstract:Supervised segmentation algorithms yield state-of-the-art results for automated anomaly detection. However, these models require voxel-wise labels which are time-consuming to draw for medical experts. An interesting alternative to voxel-wise annotations is the use of weak labels: these can be coarse or oversized annotations that are less precise, but considerably faster to create. In this work, we address the task of brain aneurysm detection by developing a fully automated, deep neural network that is trained utilizing oversized weak labels. Furthermore, since aneurysms mainly occur in specific anatomical locations, we build our model leveraging the underlying anatomy of the brain vasculature both during training and inference. We apply our model to 250 subjects (120 patients, 130 controls) who underwent Time-Of-Flight Magnetic Resonance Angiography (TOF-MRA) and presented a total of 154 aneurysms. To assess the robustness of the algorithm, we participated in a MICCAI challenge for TOF-MRA data (93 patients, 20 controls, 125 aneurysms) which allowed us to obtain results also for subjects coming from a different institution. Our network achieves an average sensitivity of 77% on our in-house data, with a mean False Positive (FP) rate of 0.72 per patient. Instead, on the challenge data, we attain a sensitivity of 59% with a mean FP rate of 1.18, ranking in 7th/14 position for detection and in 4th/11 for segmentation on the open leaderboard. When computing detection performances with respect to aneurysms' risk of rupture, we found no statistical difference between two risk groups (p = 0.12), although the sensitivity for dangerous aneurysms was higher (78%). Our approach suggests that clinically useful sensitivity can be achieved using weak labels and exploiting prior anatomical knowledge; this expands the feasibility of deep learning studies to hospitals that have limited time and data.
MBIS: Multivariate Bayesian Image Segmentation Tool
Apr 07, 2014



Abstract:We present MBIS (Multivariate Bayesian Image Segmentation tool), a clustering tool based on the mixture of multivariate normal distributions model. MBIS supports multi-channel bias field correction based on a B-spline model. A second methodological novelty is the inclusion of graph-cuts optimization for the stationary anisotropic hidden Markov random field model. Along with MBIS, we release an evaluation framework that contains three different experiments on multi-site data. We first validate the accuracy of segmentation and the estimated bias field for each channel. MBIS outperforms a widely used segmentation tool in a cross-comparison evaluation. The second experiment demonstrates the robustness of results on atlas-free segmentation of two image sets from scan-rescan protocols on 21 healthy subjects. Multivariate segmentation is more replicable than the monospectral counterpart on T1-weighted images. Finally, we provide a third experiment to illustrate how MBIS can be used in a large-scale study of tissue volume change with increasing age in 584 healthy subjects. This last result is meaningful as multivariate segmentation performs robustly without the need for prior knowledge
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge