Rastko Ciric
Department of Bioengineering, Stanford University
Differentiable programming for functional connectomics
May 31, 2022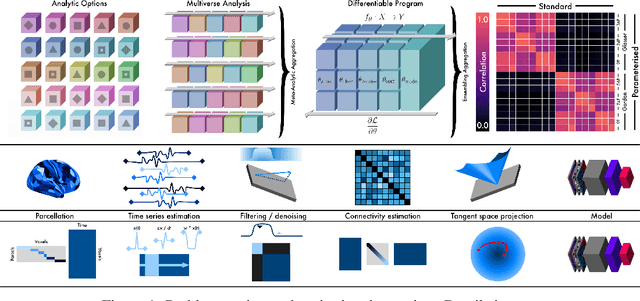
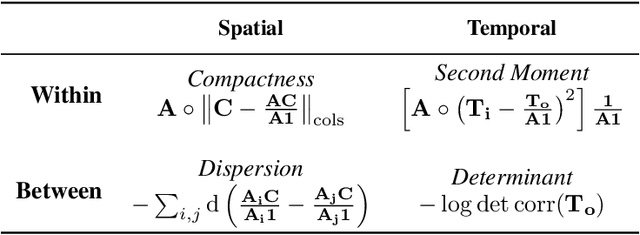
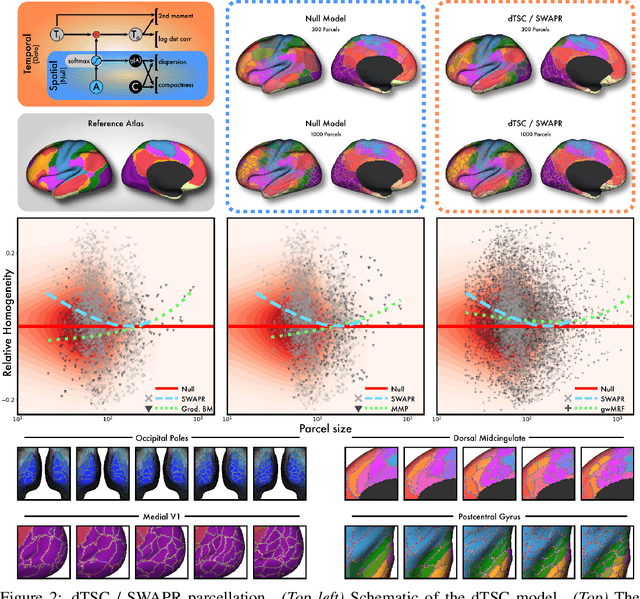
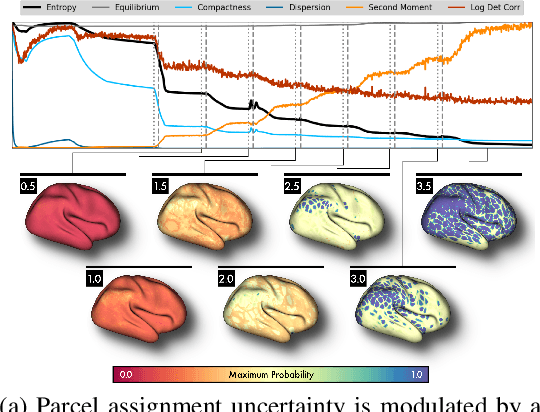
Abstract:Mapping the functional connectome has the potential to uncover key insights into brain organisation. However, existing workflows for functional connectomics are limited in their adaptability to new data, and principled workflow design is a challenging combinatorial problem. We introduce a new analytic paradigm and software toolbox that implements common operations used in functional connectomics as fully differentiable processing blocks. Under this paradigm, workflow configurations exist as reparameterisations of a differentiable functional that interpolates them. The differentiable program that we envision occupies a niche midway between traditional pipelines and end-to-end neural networks, combining the glass-box tractability and domain knowledge of the former with the amenability to optimisation of the latter. In this preliminary work, we provide a proof of concept for differentiable connectomics, demonstrating the capacity of our processing blocks both to recapitulate canonical knowledge in neuroscience and to make new discoveries in an unsupervised setting. Our differentiable modules are competitive with state-of-the-art methods in problem domains including functional parcellation, denoising, and covariance modelling. Taken together, our results and software demonstrate the promise of differentiable programming for functional connectomics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge