Hélène Lajous
Department of Radiology, Lausanne University Hospital, CIBM Center for Biomedical Imaging, Switzerland
Maximizing domain generalization in fetal brain tissue segmentation: the role of synthetic data generation, intensity clustering and real image fine-tuning
Nov 11, 2024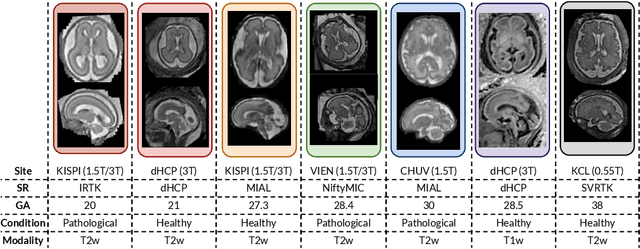
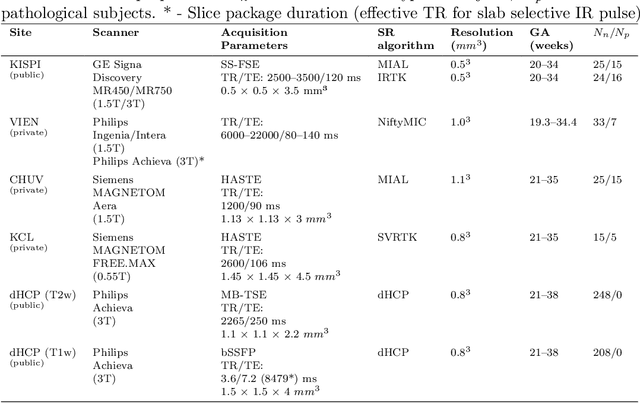
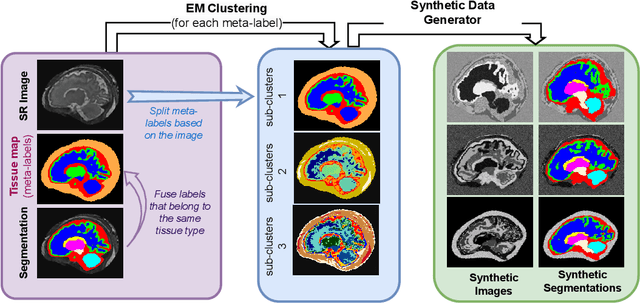
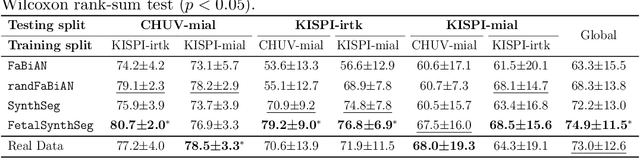
Abstract:Fetal brain tissue segmentation in magnetic resonance imaging (MRI) is a crucial tool that supports the understanding of neurodevelopment, yet it faces challenges due to the heterogeneity of data coming from different scanners and settings, and due to data scarcity. Recent approaches based on domain randomization, like SynthSeg, have shown a great potential for single source domain generalization, by simulating images with randomized contrast and image resolution from the label maps. In this work, we investigate how to maximize the out-of-domain (OOD) generalization potential of SynthSeg-based methods in fetal brain MRI. Specifically, when studying data generation, we demonstrate that the simple Gaussian mixture models used in SynthSeg enable more robust OOD generalization than physics-informed generation methods. We also investigate how intensity clustering can help create more faithful synthetic images, and observe that it is key to achieving a non-trivial OOD generalization capability when few label classes are available. Finally, by combining for the first time SynthSeg with modern fine-tuning approaches based on weight averaging, we show that fine-tuning a model pre-trained on synthetic data on a few real image-segmentation pairs in a new domain can lead to improvements in the target domain, but also in other domains. We summarize our findings as five key recommendations that we believe can guide practitioners who would like to develop SynthSeg-based approaches in other organs or modalities.
Simulation-based parameter optimization for fetal brain MRI super-resolution reconstruction
Nov 25, 2022Abstract:In utero fetal brain magnetic resonance images are inherently limited in spatial resolution due to stochastic motion of the fetus. Super-resolution reconstruction methods have become the go-to approach to compute an isotropic motion-free volume of the fetal brain from low-resolution series of 2D thick slices. Such pipelines often rely on an optimization problem with a data fidelity and a regularization term, balanced by a hyperparameter $\alpha$. The lack of ground truth images makes it difficult to adapt $\alpha$ to a given setting of interest in a quantitative manner. In this work, we propose a simulation-based approach to tune $\alpha$ for a given acquisition setting. We focus on two key aspects: the magnetic field strength (1.5T and 3T) and number of LR series used for reconstruction. Our results show that the optimal $\alpha$ significantly improves the performance compared to the default value, across two commonly used SR pipelines. Qualitative validation on clinical data confirms the importance of tuning this parameter to the setting of interest.
Self-Supervised Isotropic Superresolution Fetal Brain MRI
Nov 11, 2022Abstract:Superresolution T2-weighted fetal-brain magnetic-resonance imaging (FBMRI) traditionally relies on the availability of several orthogonal low-resolution series of 2-dimensional thick slices (volumes). In practice, only a few low-resolution volumes are acquired. Thus, optimization-based image-reconstruction methods require strong regularization using hand-crafted regularizers (e.g., TV). Yet, due to in utero fetal motion and the rapidly changing fetal brain anatomy, the acquisition of the high-resolution images that are required to train supervised learning methods is difficult. In this paper, we sidestep this difficulty by providing a proof of concept of a self-supervised single-volume superresolution framework for T2-weighted FBMRI (SAIR). We validate SAIR quantitatively in a motion-free simulated environment. Our results for different noise levels and resolution ratios suggest that SAIR is comparable to multiple-volume superresolution reconstruction methods. We also evaluate SAIR qualitatively on clinical FBMRI data. The results suggest SAIR could be incorporated into current reconstruction pipelines.
4D iterative reconstruction of brain fMRI in the moving fetus
Nov 22, 2021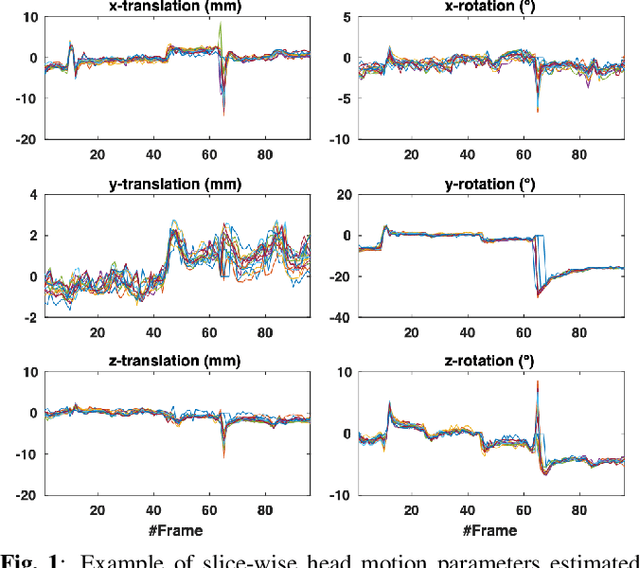
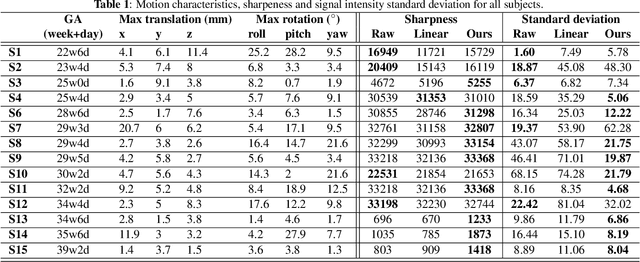
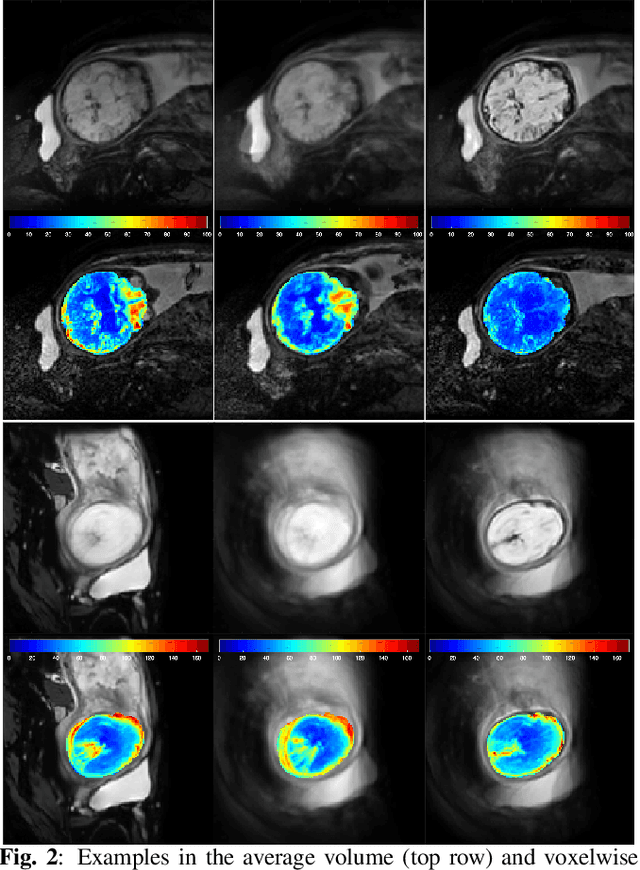
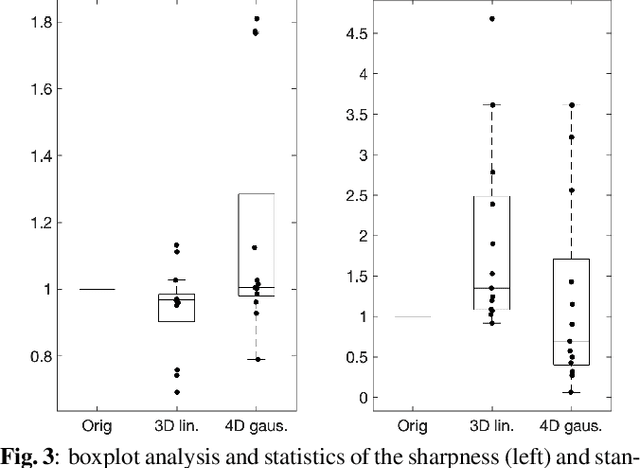
Abstract:Resting-state functional Magnetic Resonance Imaging (fMRI) is a powerful imaging technique for studying functional development of the brain in utero. However, unpredictable and excessive movement of fetuses has limited clinical application since it causes substantial signal fluctuations which can systematically alter observed patterns of functional connectivity. Previous studies have focused on the accurate estimation of the motion parameters in case of large fetal head movement and used a 3D single step interpolation approach at each timepoint to recover motion-free fMRI images. This does not guarantee that the reconstructed image corresponds to the minimum error representation of fMRI time series given the acquired data. Here, we propose a novel technique based on four dimensional iterative reconstruction of the scattered slices acquired during fetal fMRI. The accuracy of the proposed method was quantitatively evaluated on a group of real clinical fMRI fetuses. The results indicate improvements of reconstruction quality compared to the conventional 3D interpolation approach.
Synthetic magnetic resonance images for domain adaptation: Application to fetal brain tissue segmentation
Nov 08, 2021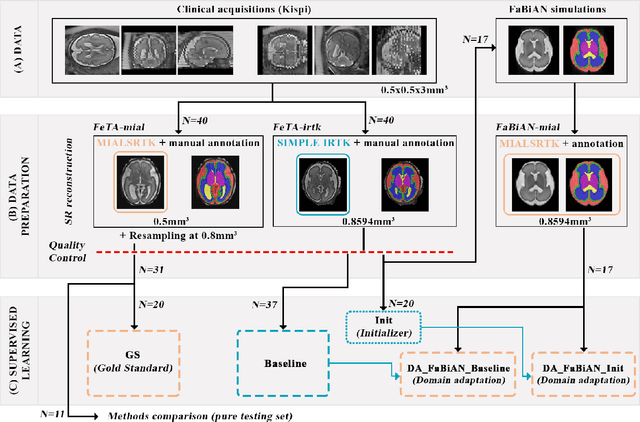
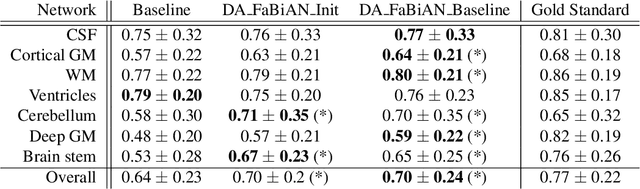
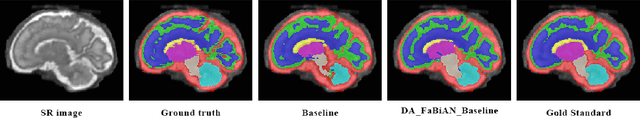
Abstract:The quantitative assessment of the developing human brain in utero is crucial to fully understand neurodevelopment. Thus, automated multi-tissue fetal brain segmentation algorithms are being developed, which in turn require annotated data to be trained. However, the available annotated fetal brain datasets are limited in number and heterogeneity, hampering domain adaptation strategies for robust segmentation. In this context, we use FaBiAN, a Fetal Brain magnetic resonance Acquisition Numerical phantom, to simulate various realistic magnetic resonance images of the fetal brain along with its class labels. We demonstrate that these multiple synthetic annotated data, generated at no cost and further reconstructed using the target super-resolution technique, can be successfully used for domain adaptation of a deep learning method that segments seven brain tissues. Overall, the accuracy of the segmentation is significantly enhanced, especially in the cortical gray matter, the white matter, the cerebellum, the deep gray matter and the brain stem.
FaBiAN: A Fetal Brain magnetic resonance Acquisition Numerical phantom
Sep 06, 2021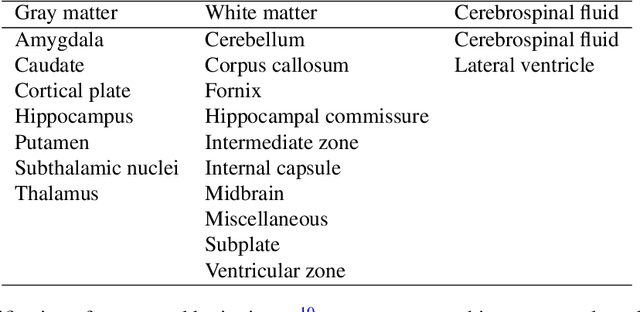
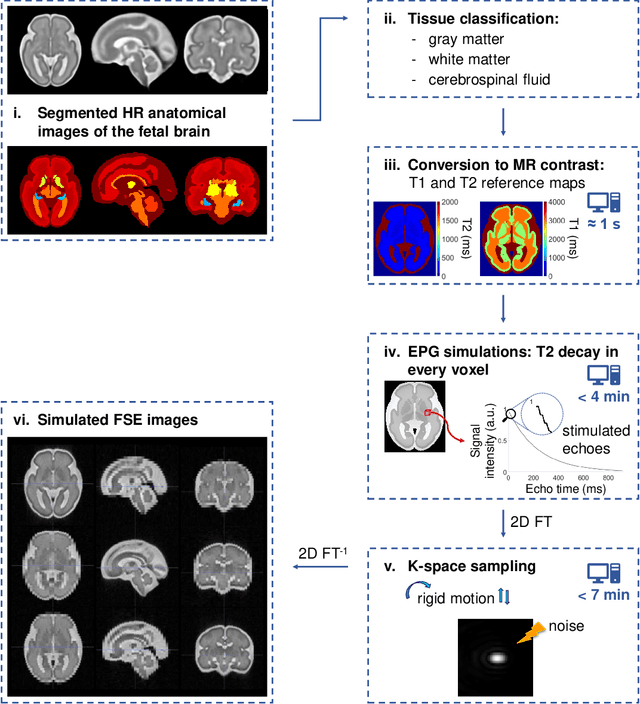

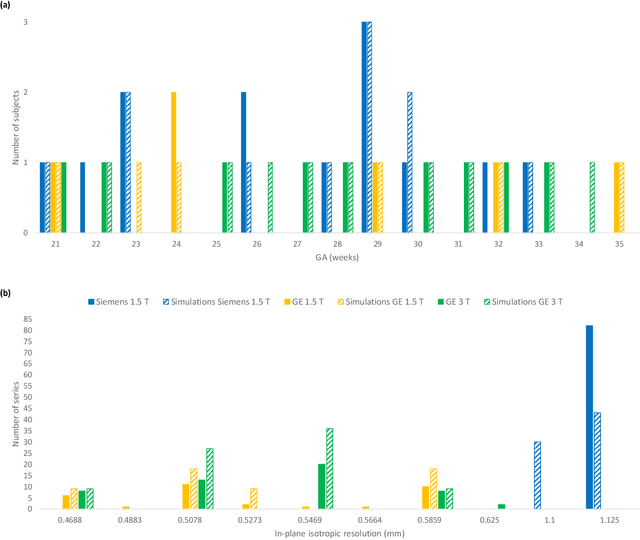
Abstract:Accurate characterization of in utero human brain maturation is critical as it involves complex and interconnected structural and functional processes that may influence health later in life. Magnetic resonance imaging is a powerful tool to investigate equivocal neurological patterns during fetal development. However, the number of acquisitions of satisfactory quality available in this cohort of sensitive subjects remains scarce, thus hindering the validation of advanced image processing techniques. Numerical phantoms can mitigate these limitations by providing a controlled environment with a known ground truth. In this work, we present FaBiAN, an open-source Fetal Brain magnetic resonance Acquisition Numerical phantom that simulates clinical T2-weighted fast spin echo sequences of the fetal brain. This unique tool is based on a general, flexible and realistic setup that includes stochastic fetal movements, thus providing images of the fetal brain throughout maturation comparable to clinical acquisitions. We demonstrate its value to evaluate the robustness and optimize the accuracy of an algorithm for super-resolution fetal brain magnetic resonance imaging from simulated motion-corrupted 2D low-resolution series as compared to a synthetic high-resolution reference volume. We also show that the images generated can complement clinical datasets to support data-intensive deep learning methods for fetal brain tissue segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge