Özgün Turgut
Multi-View Stenosis Classification Leveraging Transformer-Based Multiple-Instance Learning Using Real-World Clinical Data
Feb 02, 2026Abstract:Coronary artery stenosis is a leading cause of cardiovascular disease, diagnosed by analyzing the coronary arteries from multiple angiography views. Although numerous deep-learning models have been proposed for stenosis detection from a single angiography view, their performance heavily relies on expensive view-level annotations, which are often not readily available in hospital systems. Moreover, these models fail to capture the temporal dynamics and dependencies among multiple views, which are crucial for clinical diagnosis. To address this, we propose SegmentMIL, a transformer-based multi-view multiple-instance learning framework for patient-level stenosis classification. Trained on a real-world clinical dataset, using patient-level supervision and without any view-level annotations, SegmentMIL jointly predicts the presence of stenosis and localizes the affected anatomical region, distinguishing between the right and left coronary arteries and their respective segments. SegmentMIL obtains high performance on internal and external evaluations and outperforms both view-level models and classical MIL baselines, underscoring its potential as a clinically viable and scalable solution for coronary stenosis diagnosis. Our code is available at https://github.com/NikolaCenic/mil-stenosis.
Meta-learning Slice-to-Volume Reconstruction in Fetal Brain MRI using Implicit Neural Representations
May 14, 2025



Abstract:High-resolution slice-to-volume reconstruction (SVR) from multiple motion-corrupted low-resolution 2D slices constitutes a critical step in image-based diagnostics of moving subjects, such as fetal brain Magnetic Resonance Imaging (MRI). Existing solutions struggle with image artifacts and severe subject motion or require slice pre-alignment to achieve satisfying reconstruction performance. We propose a novel SVR method to enable fast and accurate MRI reconstruction even in cases of severe image and motion corruption. Our approach performs motion correction, outlier handling, and super-resolution reconstruction with all operations being entirely based on implicit neural representations. The model can be initialized with task-specific priors through fully self-supervised meta-learning on either simulated or real-world data. In extensive experiments including over 480 reconstructions of simulated and clinical MRI brain data from different centers, we prove the utility of our method in cases of severe subject motion and image artifacts. Our results demonstrate improvements in reconstruction quality, especially in the presence of severe motion, compared to state-of-the-art methods, and up to 50% reduction in reconstruction time.
Are foundation models useful feature extractors for electroencephalography analysis?
Feb 28, 2025Abstract:The success of foundation models in natural language processing and computer vision has motivated similar approaches for general time series analysis. While these models are effective for a variety of tasks, their applicability in medical domains with limited data remains largely unexplored. To address this, we investigate the effectiveness of foundation models in medical time series analysis involving electroencephalography (EEG). Through extensive experiments on tasks such as age prediction, seizure detection, and the classification of clinically relevant EEG events, we compare their diagnostic accuracy with that of specialised EEG models. Our analysis shows that foundation models extract meaningful EEG features, outperform specialised models even without domain adaptation, and localise task-specific biomarkers. Moreover, we demonstrate that diagnostic accuracy is substantially influenced by architectural choices such as context length. Overall, our study reveals that foundation models with general time series understanding eliminate the dependency on large domain-specific datasets, making them valuable tools for clinical practice.
Towards Generalisable Time Series Understanding Across Domains
Oct 09, 2024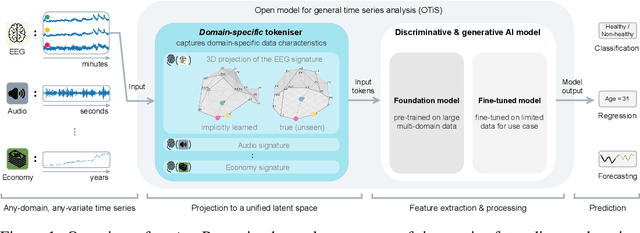
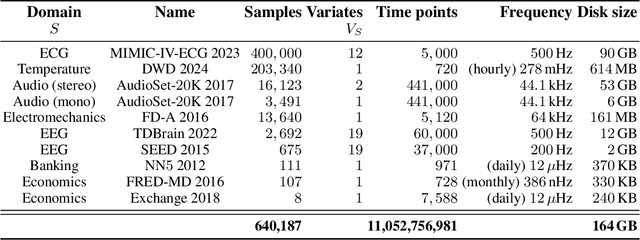
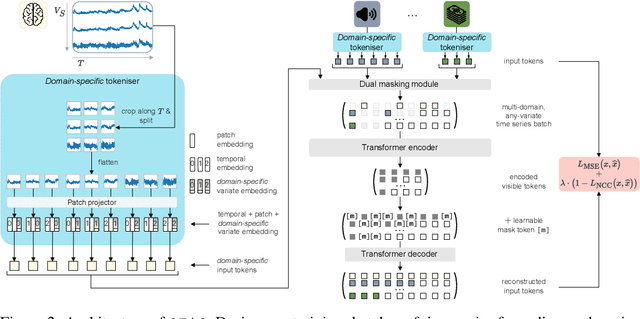
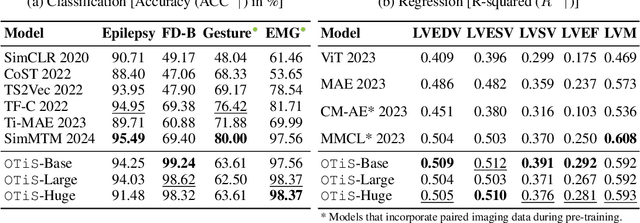
Abstract:In natural language processing and computer vision, self-supervised pre-training on large datasets unlocks foundational model capabilities across domains and tasks. However, this potential has not yet been realised in time series analysis, where existing methods disregard the heterogeneous nature of time series characteristics. Time series are prevalent in many domains, including medicine, engineering, natural sciences, and finance, but their characteristics vary significantly in terms of variate count, inter-variate relationships, temporal dynamics, and sampling frequency. This inherent heterogeneity across domains prevents effective pre-training on large time series corpora. To address this issue, we introduce OTiS, an open model for general time series analysis, that has been specifically designed to handle multi-domain heterogeneity. We propose a novel pre-training paradigm including a tokeniser with learnable domain-specific signatures, a dual masking strategy to capture temporal causality, and a normalised cross-correlation loss to model long-range dependencies. Our model is pre-trained on a large corpus of 640,187 samples and 11 billion time points spanning 8 distinct domains, enabling it to analyse time series from any (unseen) domain. In comprehensive experiments across 15 diverse applications - including classification, regression, and forecasting - OTiS showcases its ability to accurately capture domain-specific data characteristics and demonstrates its competitiveness against state-of-the-art baselines. Our code and pre-trained weights are publicly available at https://github.com/oetu/otis.
Estimating Neural Orientation Distribution Fields on High Resolution Diffusion MRI Scans
Sep 14, 2024Abstract:The Orientation Distribution Function (ODF) characterizes key brain microstructural properties and plays an important role in understanding brain structural connectivity. Recent works introduced Implicit Neural Representation (INR) based approaches to form a spatially aware continuous estimate of the ODF field and demonstrated promising results in key tasks of interest when compared to conventional discrete approaches. However, traditional INR methods face difficulties when scaling to large-scale images, such as modern ultra-high-resolution MRI scans, posing challenges in learning fine structures as well as inefficiencies in training and inference speed. In this work, we propose HashEnc, a grid-hash-encoding-based estimation of the ODF field and demonstrate its effectiveness in retaining structural and textural features. We show that HashEnc achieves a 10% enhancement in image quality while requiring 3x less computational resources than current methods. Our code can be found at https://github.com/MunzerDw/NODF-HashEnc.
Unlocking the Diagnostic Potential of ECG through Knowledge Transfer from Cardiac MRI
Aug 09, 2023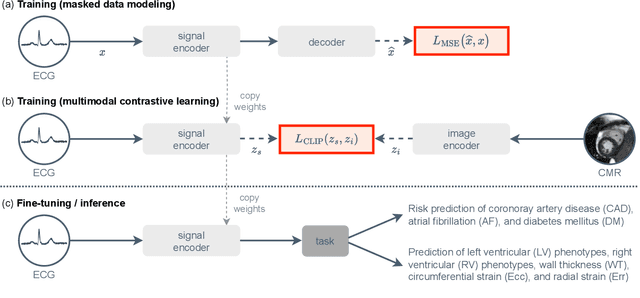
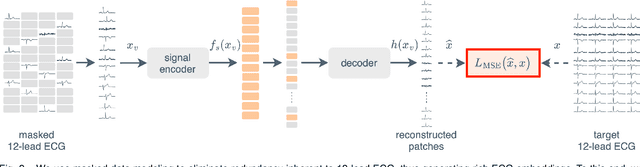
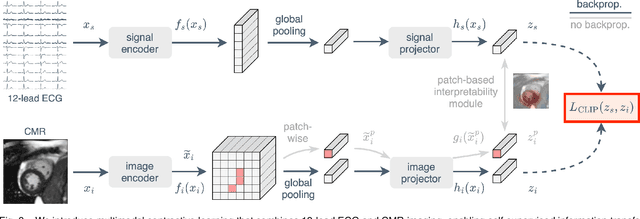
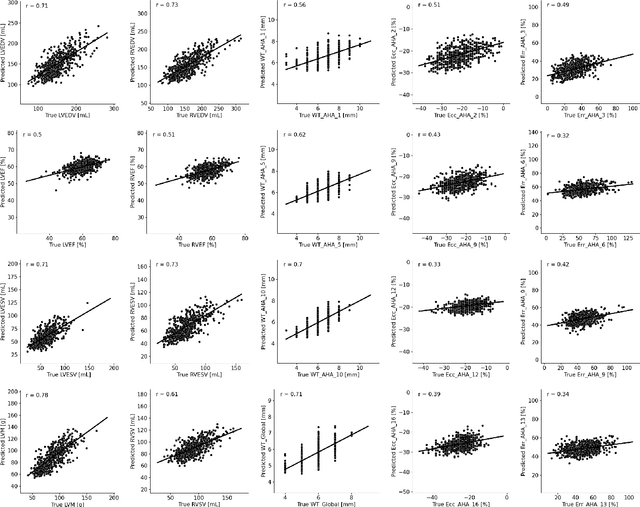
Abstract:The electrocardiogram (ECG) is a widely available diagnostic tool that allows for a cost-effective and fast assessment of the cardiovascular health. However, more detailed examination with expensive cardiac magnetic resonance (CMR) imaging is often preferred for the diagnosis of cardiovascular diseases. While providing detailed visualization of the cardiac anatomy, CMR imaging is not widely available due to long scan times and high costs. To address this issue, we propose the first self-supervised contrastive approach that transfers domain-specific information from CMR images to ECG embeddings. Our approach combines multimodal contrastive learning with masked data modeling to enable holistic cardiac screening solely from ECG data. In extensive experiments using data from 40,044 UK Biobank subjects, we demonstrate the utility and generalizability of our method. We predict the subject-specific risk of various cardiovascular diseases and determine distinct cardiac phenotypes solely from ECG data. In a qualitative analysis, we demonstrate that our learned ECG embeddings incorporate information from CMR image regions of interest. We make our entire pipeline publicly available, including the source code and pre-trained model weights.
Global k-Space Interpolation for Dynamic MRI Reconstruction using Masked Image Modeling
Jul 24, 2023


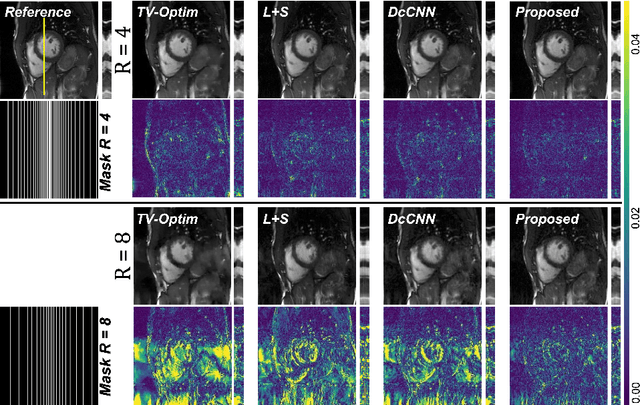
Abstract:In dynamic Magnetic Resonance Imaging (MRI), k-space is typically undersampled due to limited scan time, resulting in aliasing artifacts in the image domain. Hence, dynamic MR reconstruction requires not only modeling spatial frequency components in the x and y directions of k-space but also considering temporal redundancy. Most previous works rely on image-domain regularizers (priors) to conduct MR reconstruction. In contrast, we focus on interpolating the undersampled k-space before obtaining images with Fourier transform. In this work, we connect masked image modeling with k-space interpolation and propose a novel Transformer-based k-space Global Interpolation Network, termed k-GIN. Our k-GIN learns global dependencies among low- and high-frequency components of 2D+t k-space and uses it to interpolate unsampled data. Further, we propose a novel k-space Iterative Refinement Module (k-IRM) to enhance the high-frequency components learning. We evaluate our approach on 92 in-house 2D+t cardiac MR subjects and compare it to MR reconstruction methods with image-domain regularizers. Experiments show that our proposed k-space interpolation method quantitatively and qualitatively outperforms baseline methods. Importantly, the proposed approach achieves substantially higher robustness and generalizability in cases of highly-undersampled MR data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge