Paul Hager
on behalf of the PINNACLE consortium
Towards Cardiac MRI Foundation Models: Comprehensive Visual-Tabular Representations for Whole-Heart Assessment and Beyond
Apr 18, 2025Abstract:Cardiac magnetic resonance imaging is the gold standard for non-invasive cardiac assessment, offering rich spatio-temporal views of the cardiac anatomy and physiology. Patient-level health factors, such as demographics, metabolic, and lifestyle, are known to substantially influence cardiovascular health and disease risk, yet remain uncaptured by CMR alone. To holistically understand cardiac health and to enable the best possible interpretation of an individual's disease risk, CMR and patient-level factors must be jointly exploited within an integrated framework. Recent multi-modal approaches have begun to bridge this gap, yet they often rely on limited spatio-temporal data and focus on isolated clinical tasks, thereby hindering the development of a comprehensive representation for cardiac health evaluation. To overcome these limitations, we introduce ViTa, a step toward foundation models that delivers a comprehensive representation of the heart and a precise interpretation of individual disease risk. Leveraging data from 42,000 UK Biobank participants, ViTa integrates 3D+T cine stacks from short-axis and long-axis views, enabling a complete capture of the cardiac cycle. These imaging data are then fused with detailed tabular patient-level factors, enabling context-aware insights. This multi-modal paradigm supports a wide spectrum of downstream tasks, including cardiac phenotype and physiological feature prediction, segmentation, and classification of cardiac and metabolic diseases within a single unified framework. By learning a shared latent representation that bridges rich imaging features and patient context, ViTa moves beyond traditional, task-specific models toward a universal, patient-specific understanding of cardiac health, highlighting its potential to advance clinical utility and scalability in cardiac analysis.
Self-Supervised Radiograph Anatomical Region Classification -- How Clean Is Your Real-World Data?
Dec 20, 2024Abstract:Modern deep learning-based clinical imaging workflows rely on accurate labels of the examined anatomical region. Knowing the anatomical region is required to select applicable downstream models and to effectively generate cohorts of high quality data for future medical and machine learning research efforts. However, this information may not be available in externally sourced data or generally contain data entry errors. To address this problem, we show the effectiveness of self-supervised methods such as SimCLR and BYOL as well as supervised contrastive deep learning methods in assigning one of 14 anatomical region classes in our in-house dataset of 48,434 skeletal radiographs. We achieve a strong linear evaluation accuracy of 96.6% with a single model and 97.7% using an ensemble approach. Furthermore, only a few labeled instances (1% of the training set) suffice to achieve an accuracy of 92.2%, enabling usage in low-label and thus low-resource scenarios. Our model can be used to correct data entry mistakes: a follow-up analysis of the test set errors of our best-performing single model by an expert radiologist identified 35% incorrect labels and 11% out-of-domain images. When accounted for, the radiograph anatomical region labelling performance increased -- without and with an ensemble, respectively -- to a theoretical accuracy of 98.0% and 98.8%.
Specialist vision-language models for clinical ophthalmology
Jul 11, 2024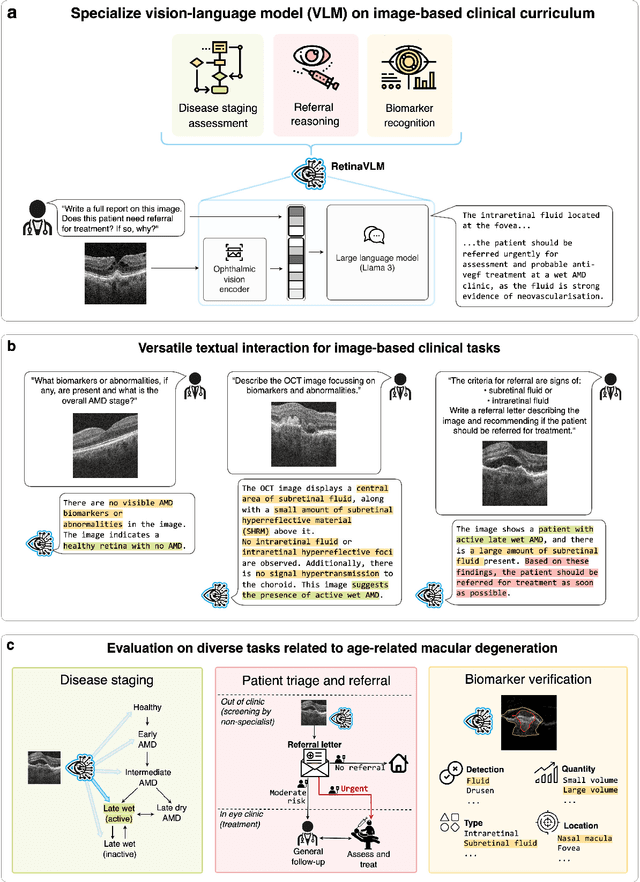

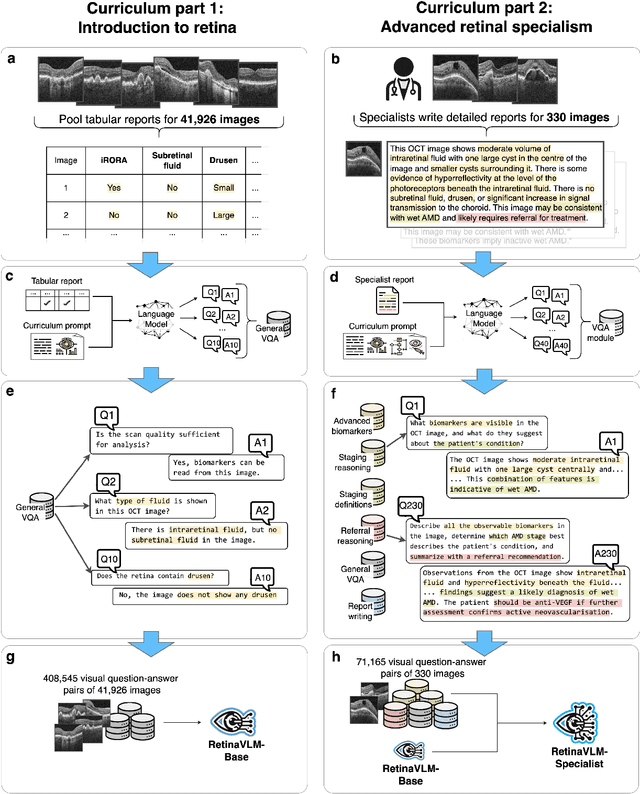

Abstract:Clinicians spend a significant amount of time reviewing medical images and transcribing their findings regarding patient diagnosis, referral and treatment in text form. Vision-language models (VLMs), which automatically interpret images and summarize their findings as text, have enormous potential to alleviate clinical workloads and increase patient access to high-quality medical care. While foundational models have stirred considerable interest in the medical community, it is unclear whether their general capabilities translate to real-world clinical utility. In this work, we show that foundation VLMs markedly underperform compared to practicing ophthalmologists on specialist tasks crucial to the care of patients with age-related macular degeneration (AMD). To address this, we initially identified the essential capabilities required for image-based clinical decision-making, and then developed a curriculum to selectively train VLMs in these skills. The resulting model, RetinaVLM, can be instructed to write reports that significantly outperform those written by leading foundation medical VLMs in disease staging (F1 score of 0.63 vs. 0.11) and patient referral (0.67 vs. 0.39), and approaches the diagnostic performance of junior ophthalmologists (who achieve 0.77 and 0.78 on the respective tasks). Furthermore, in a reader study involving two senior ophthalmologists with up to 32 years of experience, RetinaVLM's reports were found to be similarly correct (78.6% vs. 82.1%) and complete (both 78.6%) as reports written by junior ophthalmologists with up to 10 years of experience. These results demonstrate that our curriculum-based approach provides a blueprint for specializing generalist foundation medical VLMs to handle real-world clinical tasks.
Spatiotemporal Representation Learning for Short and Long Medical Image Time Series
Mar 12, 2024Abstract:Analyzing temporal developments is crucial for the accurate prognosis of many medical conditions. Temporal changes that occur over short time scales are key to assessing the health of physiological functions, such as the cardiac cycle. Moreover, tracking longer term developments that occur over months or years in evolving processes, such as age-related macular degeneration (AMD), is essential for accurate prognosis. Despite the importance of both short and long term analysis to clinical decision making, they remain understudied in medical deep learning. State of the art methods for spatiotemporal representation learning, developed for short natural videos, prioritize the detection of temporal constants rather than temporal developments. Moreover, they do not account for varying time intervals between acquisitions, which are essential for contextualizing observed changes. To address these issues, we propose two approaches. First, we combine clip-level contrastive learning with a novel temporal embedding to adapt to irregular time series. Second, we propose masking and predicting latent frame representations of the temporal sequence. Our two approaches outperform all prior methods on temporally-dependent tasks including cardiac output estimation and three prognostic AMD tasks. Overall, this enables the automated analysis of temporal patterns which are typically overlooked in applications of deep learning to medicine.
Propagation and Attribution of Uncertainty in Medical Imaging Pipelines
Sep 28, 2023Abstract:Uncertainty estimation, which provides a means of building explainable neural networks for medical imaging applications, have mostly been studied for single deep learning models that focus on a specific task. In this paper, we propose a method to propagate uncertainty through cascades of deep learning models in medical imaging pipelines. This allows us to aggregate the uncertainty in later stages of the pipeline and to obtain a joint uncertainty measure for the predictions of later models. Additionally, we can separately report contributions of the aleatoric, data-based, uncertainty of every component in the pipeline. We demonstrate the utility of our method on a realistic imaging pipeline that reconstructs undersampled brain and knee magnetic resonance (MR) images and subsequently predicts quantitative information from the images, such as the brain volume, or knee side or patient's sex. We quantitatively show that the propagated uncertainty is correlated with input uncertainty and compare the proportions of contributions of pipeline stages to the joint uncertainty measure.
Unlocking the Diagnostic Potential of ECG through Knowledge Transfer from Cardiac MRI
Aug 09, 2023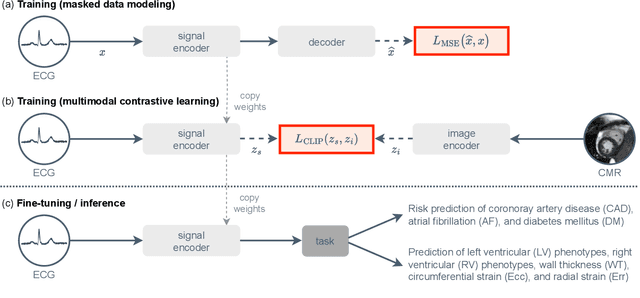
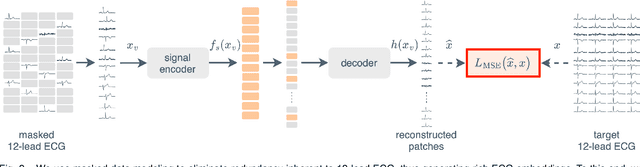
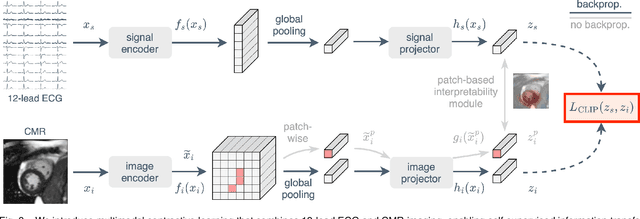
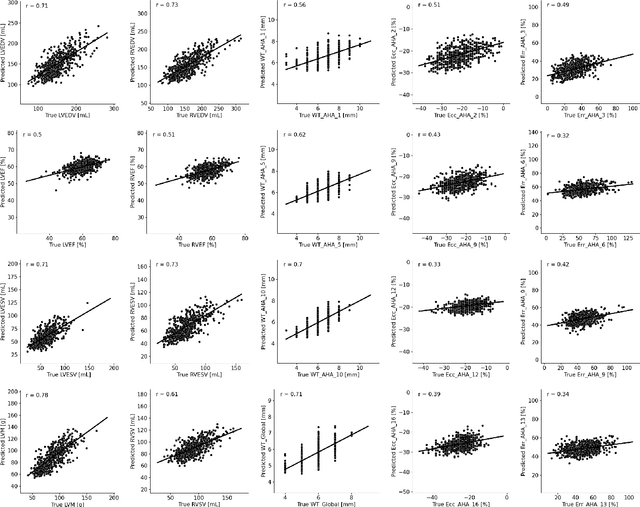
Abstract:The electrocardiogram (ECG) is a widely available diagnostic tool that allows for a cost-effective and fast assessment of the cardiovascular health. However, more detailed examination with expensive cardiac magnetic resonance (CMR) imaging is often preferred for the diagnosis of cardiovascular diseases. While providing detailed visualization of the cardiac anatomy, CMR imaging is not widely available due to long scan times and high costs. To address this issue, we propose the first self-supervised contrastive approach that transfers domain-specific information from CMR images to ECG embeddings. Our approach combines multimodal contrastive learning with masked data modeling to enable holistic cardiac screening solely from ECG data. In extensive experiments using data from 40,044 UK Biobank subjects, we demonstrate the utility and generalizability of our method. We predict the subject-specific risk of various cardiovascular diseases and determine distinct cardiac phenotypes solely from ECG data. In a qualitative analysis, we demonstrate that our learned ECG embeddings incorporate information from CMR image regions of interest. We make our entire pipeline publicly available, including the source code and pre-trained model weights.
Best of Both Worlds: Multimodal Contrastive Learning with Tabular and Imaging Data
Mar 30, 2023Abstract:Medical datasets and especially biobanks, often contain extensive tabular data with rich clinical information in addition to images. In practice, clinicians typically have less data, both in terms of diversity and scale, but still wish to deploy deep learning solutions. Combined with increasing medical dataset sizes and expensive annotation costs, the necessity for unsupervised methods that can pretrain multimodally and predict unimodally has risen. To address these needs, we propose the first self-supervised contrastive learning framework that takes advantage of images and tabular data to train unimodal encoders. Our solution combines SimCLR and SCARF, two leading contrastive learning strategies, and is simple and effective. In our experiments, we demonstrate the strength of our framework by predicting risks of myocardial infarction and coronary artery disease (CAD) using cardiac MR images and 120 clinical features from 40,000 UK Biobank subjects. Furthermore, we show the generalizability of our approach to natural images using the DVM car advertisement dataset. We take advantage of the high interpretability of tabular data and through attribution and ablation experiments find that morphometric tabular features, describing size and shape, have outsized importance during the contrastive learning process and improve the quality of the learned embeddings. Finally, we introduce a novel form of supervised contrastive learning, label as a feature (LaaF), by appending the ground truth label as a tabular feature during multimodal pretraining, outperforming all supervised contrastive baselines.
Reinforced optimal control
Nov 24, 2020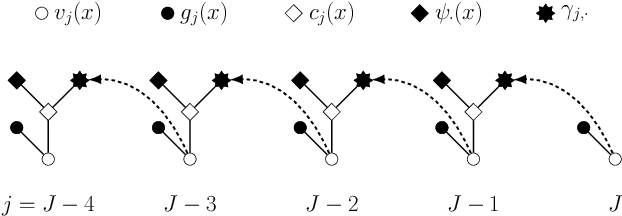
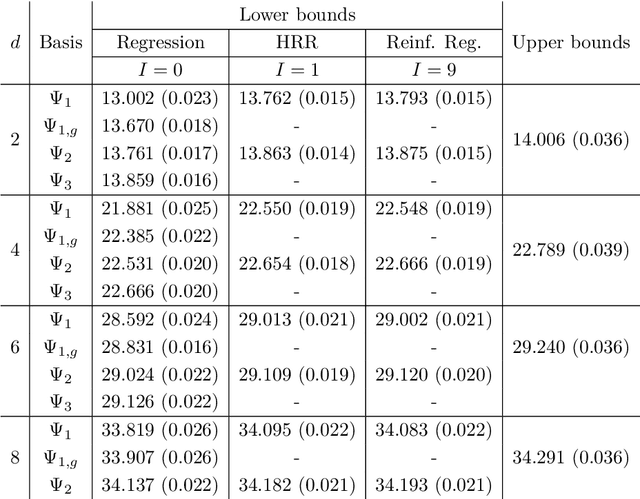
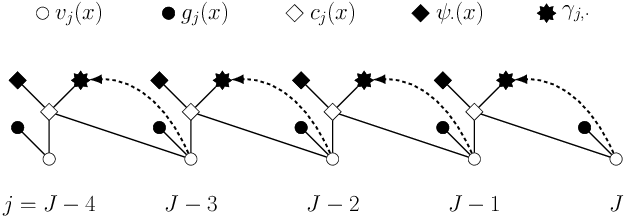
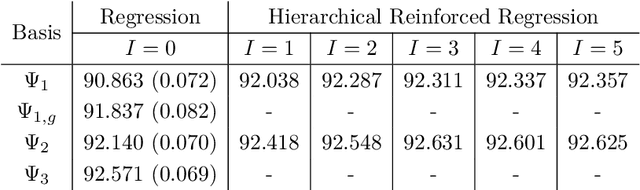
Abstract:Least squares Monte Carlo methods are a popular numerical approximation method for solving stochastic control problems. Based on dynamic programming, their key feature is the approximation of the conditional expectation of future rewards by linear least squares regression. Hence, the choice of basis functions is crucial for the accuracy of the method. Earlier work by some of us [Belomestny, Schoenmakers, Spokoiny, Zharkynbay. Commun.~Math.~Sci., 18(1):109-121, 2020] proposes to \emph{reinforce} the basis functions in the case of optimal stopping problems by already computed value functions for later times, thereby considerably improving the accuracy with limited additional computational cost. We extend the reinforced regression method to a general class of stochastic control problems, while considerably improving the method's efficiency, as demonstrated by substantial numerical examples as well as theoretical analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge