Sophie Riedl
on behalf of the PINNACLE consortium
Specialist vision-language models for clinical ophthalmology
Jul 11, 2024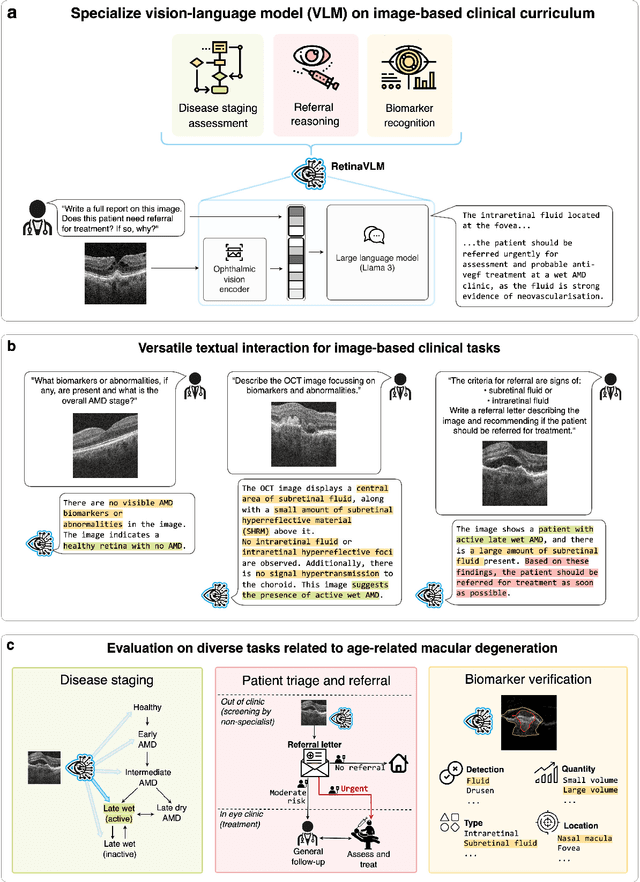

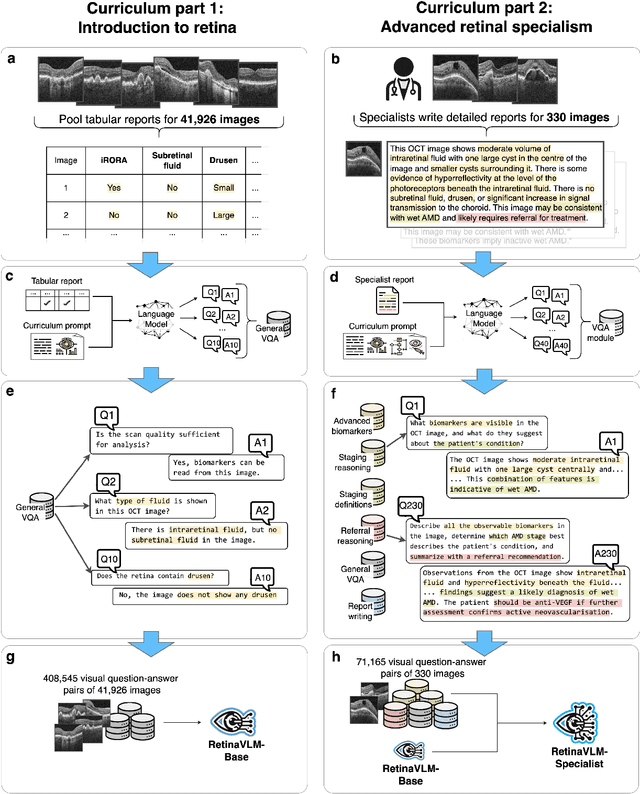

Abstract:Clinicians spend a significant amount of time reviewing medical images and transcribing their findings regarding patient diagnosis, referral and treatment in text form. Vision-language models (VLMs), which automatically interpret images and summarize their findings as text, have enormous potential to alleviate clinical workloads and increase patient access to high-quality medical care. While foundational models have stirred considerable interest in the medical community, it is unclear whether their general capabilities translate to real-world clinical utility. In this work, we show that foundation VLMs markedly underperform compared to practicing ophthalmologists on specialist tasks crucial to the care of patients with age-related macular degeneration (AMD). To address this, we initially identified the essential capabilities required for image-based clinical decision-making, and then developed a curriculum to selectively train VLMs in these skills. The resulting model, RetinaVLM, can be instructed to write reports that significantly outperform those written by leading foundation medical VLMs in disease staging (F1 score of 0.63 vs. 0.11) and patient referral (0.67 vs. 0.39), and approaches the diagnostic performance of junior ophthalmologists (who achieve 0.77 and 0.78 on the respective tasks). Furthermore, in a reader study involving two senior ophthalmologists with up to 32 years of experience, RetinaVLM's reports were found to be similarly correct (78.6% vs. 82.1%) and complete (both 78.6%) as reports written by junior ophthalmologists with up to 10 years of experience. These results demonstrate that our curriculum-based approach provides a blueprint for specializing generalist foundation medical VLMs to handle real-world clinical tasks.
3DTINC: Time-Equivariant Non-Contrastive Learning for Predicting Disease Progression from Longitudinal OCTs
Dec 28, 2023



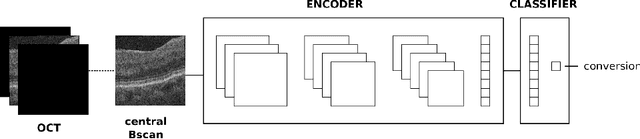
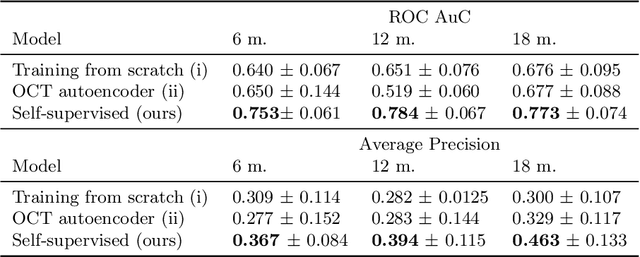
Abstract:Self-supervised learning (SSL) has emerged as a powerful technique for improving the efficiency and effectiveness of deep learning models. Contrastive methods are a prominent family of SSL that extract similar representations of two augmented views of an image while pushing away others in the representation space as negatives. However, the state-of-the-art contrastive methods require large batch sizes and augmentations designed for natural images that are impractical for 3D medical images. To address these limitations, we propose a new longitudinal SSL method, 3DTINC, based on non-contrastive learning. It is designed to learn perturbation-invariant features for 3D optical coherence tomography (OCT) volumes, using augmentations specifically designed for OCT. We introduce a new non-contrastive similarity loss term that learns temporal information implicitly from intra-patient scans acquired at different times. Our experiments show that this temporal information is crucial for predicting progression of retinal diseases, such as age-related macular degeneration (AMD). After pretraining with 3DTINC, we evaluated the learned representations and the prognostic models on two large-scale longitudinal datasets of retinal OCTs where we predict the conversion to wet-AMD within a six months interval. Our results demonstrate that each component of our contributions is crucial for learning meaningful representations useful in predicting disease progression from longitudinal volumetric scans.
Pretrained Deep 2.5D Models for Efficient Predictive Modeling from Retinal OCT
Jul 25, 2023Abstract:In the field of medical imaging, 3D deep learning models play a crucial role in building powerful predictive models of disease progression. However, the size of these models presents significant challenges, both in terms of computational resources and data requirements. Moreover, achieving high-quality pretraining of 3D models proves to be even more challenging. To address these issues, hybrid 2.5D approaches provide an effective solution for utilizing 3D volumetric data efficiently using 2D models. Combining 2D and 3D techniques offers a promising avenue for optimizing performance while minimizing memory requirements. In this paper, we explore 2.5D architectures based on a combination of convolutional neural networks (CNNs), long short-term memory (LSTM), and Transformers. In addition, leveraging the benefits of recent non-contrastive pretraining approaches in 2D, we enhanced the performance and data efficiency of 2.5D techniques even further. We demonstrate the effectiveness of architectures and associated pretraining on a task of predicting progression to wet age-related macular degeneration (AMD) within a six-month period on two large longitudinal OCT datasets.
Morph-SSL: Self-Supervision with Longitudinal Morphing to Predict AMD Progression from OCT
Apr 17, 2023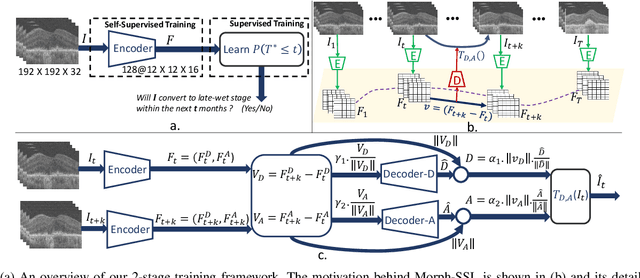
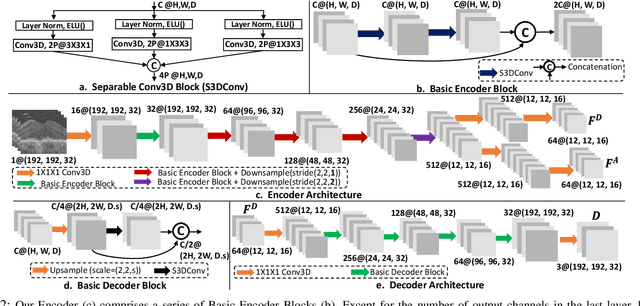

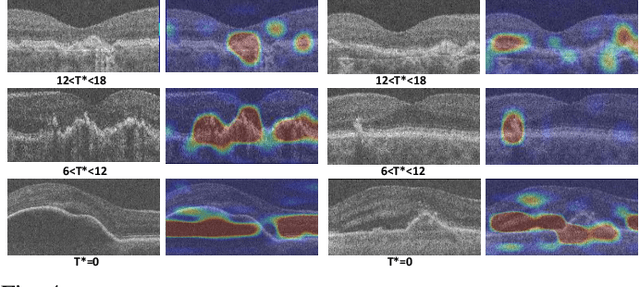
Abstract:The lack of reliable biomarkers makes predicting the conversion from intermediate to neovascular age-related macular degeneration (iAMD, nAMD) a challenging task. We develop a Deep Learning (DL) model to predict the future risk of conversion of an eye from iAMD to nAMD from its current OCT scan. Although eye clinics generate vast amounts of longitudinal OCT scans to monitor AMD progression, only a small subset can be manually labeled for supervised DL. To address this issue, we propose Morph-SSL, a novel Self-supervised Learning (SSL) method for longitudinal data. It uses pairs of unlabelled OCT scans from different visits and involves morphing the scan from the previous visit to the next. The Decoder predicts the transformation for morphing and ensures a smooth feature manifold that can generate intermediate scans between visits through linear interpolation. Next, the Morph-SSL trained features are input to a Classifier which is trained in a supervised manner to model the cumulative probability distribution of the time to conversion with a sigmoidal function. Morph-SSL was trained on unlabelled scans of 399 eyes (3570 visits). The Classifier was evaluated with a five-fold cross-validation on 2418 scans from 343 eyes with clinical labels of the conversion date. The Morph-SSL features achieved an AUC of 0.766 in predicting the conversion to nAMD within the next 6 months, outperforming the same network when trained end-to-end from scratch or pre-trained with popular SSL methods. Automated prediction of the future risk of nAMD onset can enable timely treatment and individualized AMD management.
Clustering disease trajectories in contrastive feature space for biomarker discovery in age-related macular degeneration
Jan 11, 2023Abstract:Age-related macular degeneration (AMD) is the leading cause of blindness in the elderly. Despite this, the exact dynamics of disease progression are poorly understood. There is a clear need for imaging biomarkers in retinal optical coherence tomography (OCT) that aid the diagnosis, prognosis and management of AMD. However, current grading systems, which coarsely group disease stage into broad categories describing early and intermediate AMD, have very limited prognostic value for the conversion to late AMD. In this paper, we are the first to analyse disease progression as clustered trajectories in a self-supervised feature space. Our method first pretrains an encoder with contrastive learning to project images from longitudinal time series to points in feature space. This enables the creation of disease trajectories, which are then denoised, partitioned and grouped into clusters. These clusters, found in two datasets containing time series of 7,912 patients imaged over eight years, were correlated with known OCT biomarkers. This reinforced efforts by four expert ophthalmologists to investigate clusters, during a clinical comparison and interpretation task, as candidates for time-dependent biomarkers that describe progression of AMD.
Metadata-enhanced contrastive learning from retinal optical coherence tomography images
Aug 04, 2022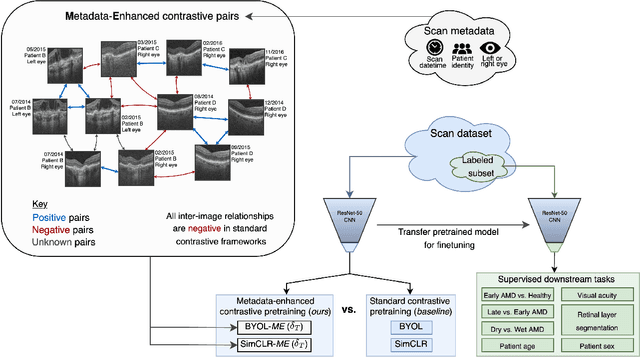
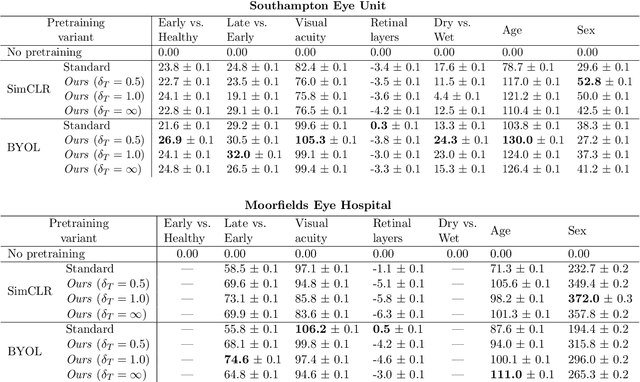
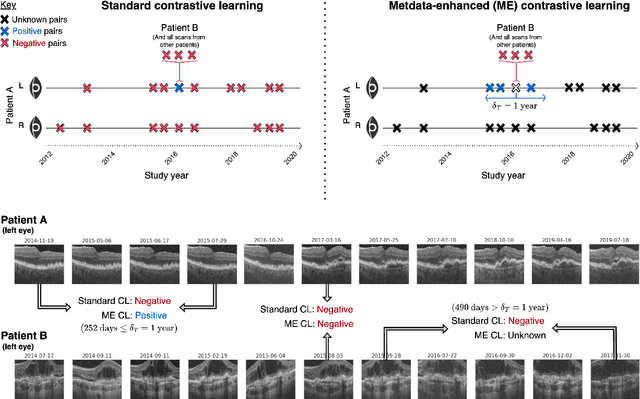
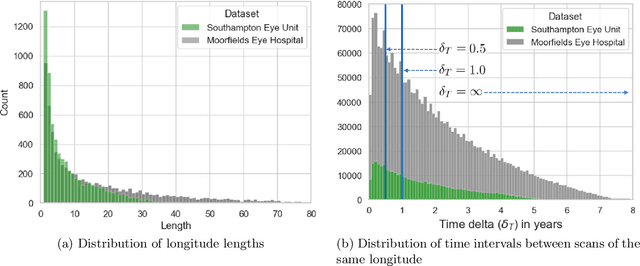
Abstract:Supervised deep learning algorithms hold great potential to automate screening, monitoring and grading of medical images. However, training performant models has typically required vast quantities of labelled data, which is scarcely available in the medical domain. Self-supervised contrastive frameworks relax this dependency by first learning from unlabelled images. In this work we show that pretraining with two contrastive methods, SimCLR and BYOL, improves the utility of deep learning with regard to the clinical assessment of age-related macular degeneration (AMD). In experiments using two large clinical datasets containing 170,427 optical coherence tomography (OCT) images of 7,912 patients, we evaluate benefits attributed to pretraining across seven downstream tasks ranging from AMD stage and type classification to prediction of functional endpoints to segmentation of retinal layers, finding performance significantly increased in six out of seven tasks with fewer labels. However, standard contrastive frameworks have two known weaknesses that are detrimental to pretraining in the medical domain. Several of the image transformations used to create positive contrastive pairs are not applicable to greyscale medical scans. Furthermore, medical images often depict the same anatomical region and disease severity, resulting in numerous misleading negative pairs. To address these issues we develop a novel metadata-enhanced approach that exploits the rich set of inherently available patient information. To this end we employ records for patient identity, eye position (i.e. left or right) and time series data to indicate the typically unknowable set of inter-image contrastive relationships. By leveraging this often neglected information our metadata-enhanced contrastive pretraining leads to further benefits and outperforms conventional contrastive methods in five out of seven downstream tasks.
SD-LayerNet: Semi-supervised retinal layer segmentation in OCT using disentangled representation with anatomical priors
Jul 01, 2022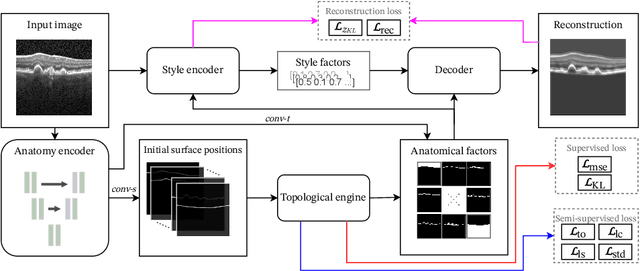
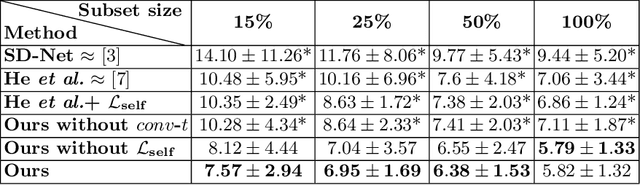
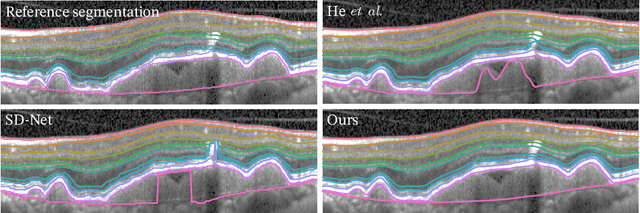
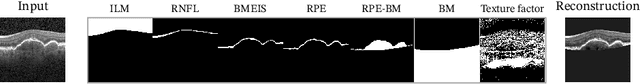
Abstract:Optical coherence tomography (OCT) is a non-invasive 3D modality widely used in ophthalmology for imaging the retina. Achieving automated, anatomically coherent retinal layer segmentation on OCT is important for the detection and monitoring of different retinal diseases, like Age-related Macular Disease (AMD) or Diabetic Retinopathy. However, the majority of state-of-the-art layer segmentation methods are based on purely supervised deep-learning, requiring a large amount of pixel-level annotated data that is expensive and hard to obtain. With this in mind, we introduce a semi-supervised paradigm into the retinal layer segmentation task that makes use of the information present in large-scale unlabeled datasets as well as anatomical priors. In particular, a novel fully differentiable approach is used for converting surface position regression into a pixel-wise structured segmentation, allowing to use both 1D surface and 2D layer representations in a coupled fashion to train the model. In particular, these 2D segmentations are used as anatomical factors that, together with learned style factors, compose disentangled representations used for reconstructing the input image. In parallel, we propose a set of anatomical priors to improve network training when a limited amount of labeled data is available. We demonstrate on the real-world dataset of scans with intermediate and wet-AMD that our method outperforms state-of-the-art when using our full training set, but more importantly largely exceeds state-of-the-art when it is trained with a fraction of the labeled data.
TINC: Temporally Informed Non-Contrastive Learning for Disease Progression Modeling in Retinal OCT Volumes
Jun 30, 2022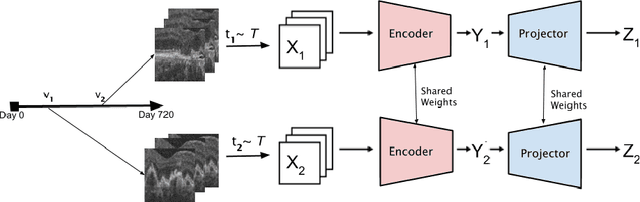
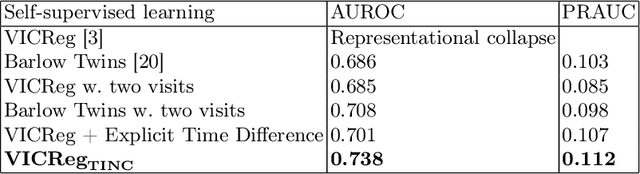
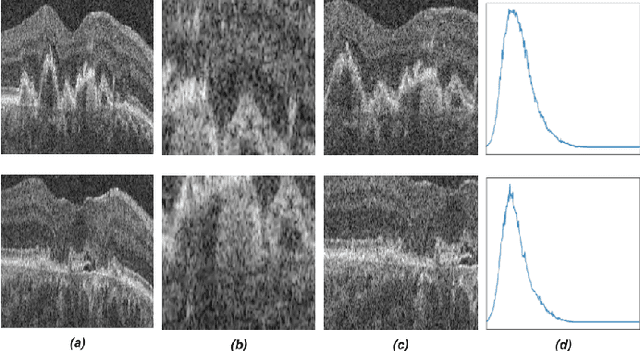

Abstract:Recent contrastive learning methods achieved state-of-the-art in low label regimes. However, the training requires large batch sizes and heavy augmentations to create multiple views of an image. With non-contrastive methods, the negatives are implicitly incorporated in the loss, allowing different images and modalities as pairs. Although the meta-information (i.e., age, sex) in medical imaging is abundant, the annotations are noisy and prone to class imbalance. In this work, we exploited already existing temporal information (different visits from a patient) in a longitudinal optical coherence tomography (OCT) dataset using temporally informed non-contrastive loss (TINC) without increasing complexity and need for negative pairs. Moreover, our novel pair-forming scheme can avoid heavy augmentations and implicitly incorporates the temporal information in the pairs. Finally, these representations learned from the pretraining are more successful in predicting disease progression where the temporal information is crucial for the downstream task. More specifically, our model outperforms existing models in predicting the risk of conversion within a time frame from intermediate age-related macular degeneration (AMD) to the late wet-AMD stage.
Modeling Disease Progression In Retinal OCTs With Longitudinal Self-Supervised Learning
Oct 24, 2019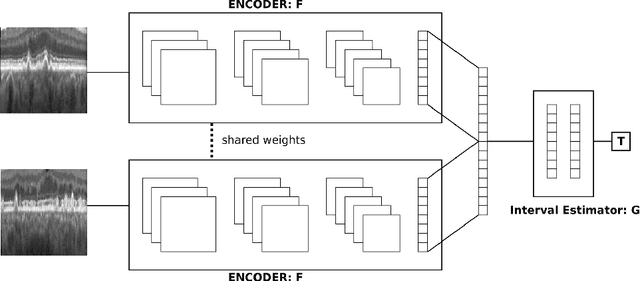
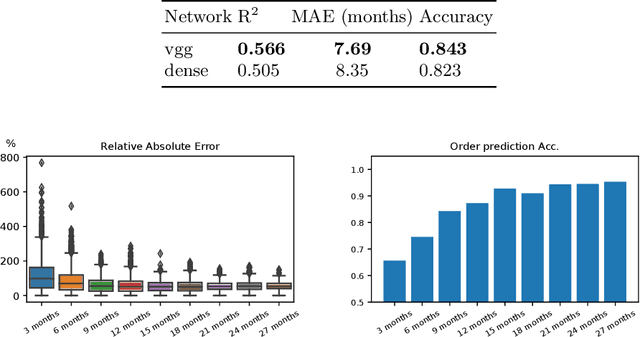
Abstract:Longitudinal imaging is capable of capturing the static ana\-to\-mi\-cal structures and the dynamic changes of the morphology resulting from aging or disease progression. Self-supervised learning allows to learn new representation from available large unlabelled data without any expert knowledge. We propose a deep learning self-supervised approach to model disease progression from longitudinal retinal optical coherence tomography (OCT). Our self-supervised model takes benefit from a generic time-related task, by learning to estimate the time interval between pairs of scans acquired from the same patient. This task is (i) easy to implement, (ii) allows to use irregularly sampled data, (iii) is tolerant to poor registration, and (iv) does not rely on additional annotations. This novel method learns a representation that focuses on progression specific information only, which can be transferred to other types of longitudinal problems. We transfer the learnt representation to a clinically highly relevant task of predicting the onset of an advanced stage of age-related macular degeneration within a given time interval based on a single OCT scan. The boost in prediction accuracy, in comparison to a network learned from scratch or transferred from traditional tasks, demonstrates that our pretrained self-supervised representation learns a clinically meaningful information.
An amplified-target loss approach for photoreceptor layer segmentation in pathological OCT scans
Aug 02, 2019



Abstract:Segmenting anatomical structures such as the photoreceptor layer in retinal optical coherence tomography (OCT) scans is challenging in pathological scenarios. Supervised deep learning models trained with standard loss functions are usually able to characterize only the most common disease appeareance from a training set, resulting in suboptimal performance and poor generalization when dealing with unseen lesions. In this paper we propose to overcome this limitation by means of an augmented target loss function framework. We introduce a novel amplified-target loss that explicitly penalizes errors within the central area of the input images, based on the observation that most of the challenging disease appeareance is usually located in this area. We experimentally validated our approach using a data set with OCT scans of patients with macular diseases. We observe increased performance compared to the models that use only the standard losses. Our proposed loss function strongly supports the segmentation model to better distinguish photoreceptors in highly pathological scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge