Guilherme Aresta
Automatic detection and prediction of nAMD activity change in retinal OCT using Siamese networks and Wasserstein Distance for ordinality
Jan 24, 2025Abstract:Neovascular age-related macular degeneration (nAMD) is a leading cause of vision loss among older adults, where disease activity detection and progression prediction are critical for nAMD management in terms of timely drug administration and improving patient outcomes. Recent advancements in deep learning offer a promising solution for predicting changes in AMD from optical coherence tomography (OCT) retinal volumes. In this work, we proposed deep learning models for the two tasks of the public MARIO Challenge at MICCAI 2024, designed to detect and forecast changes in nAMD severity with longitudinal retinal OCT. For the first task, we employ a Vision Transformer (ViT) based Siamese Network to detect changes in AMD severity by comparing scan embeddings of a patient from different time points. To train a model to forecast the change after 3 months, we exploit, for the first time, an Earth Mover (Wasserstein) Distance-based loss to harness the ordinal relation within the severity change classes. Both models ranked high on the preliminary leaderboard, demonstrating that their predictive capabilities could facilitate nAMD treatment management.
RRWNet: Recursive Refinement Network for Effective Retinal Artery/Vein Segmentation and Classification
Feb 05, 2024



Abstract:The caliber and configuration of retinal blood vessels serve as important biomarkers for various diseases and medical conditions. A thorough analysis of the retinal vasculature requires the segmentation of blood vessels and their classification into arteries and veins, which is typically performed on color fundus images obtained by retinography, a widely used imaging technique. Nonetheless, manually performing these tasks is labor-intensive and prone to human error. Various automated methods have been proposed to address this problem. However, the current state of art in artery/vein segmentation and classification faces challenges due to manifest classification errors that affect the topological consistency of segmentation maps. This study presents an innovative end-to-end framework, RRWNet, designed to recursively refine semantic segmentation maps and correct manifest classification errors. The framework consists of a fully convolutional neural network with a Base subnetwork that generates base segmentation maps from input images, and a Recursive Refinement subnetwork that iteratively and recursively improves these maps. Evaluation on public datasets demonstrates the state-of-the-art performance of the proposed method, yielding more topologically consistent segmentation maps with fewer manifest classification errors than existing approaches. In addition, the Recursive Refinement module proves effective in post-processing segmentation maps from other methods, automatically correcting classification errors and improving topological consistency. The model code, weights, and predictions are publicly available at https://github.com/j-morano/rrwnet.
Deep Multimodal Fusion of Data with Heterogeneous Dimensionality via Projective Networks
Feb 02, 2024



Abstract:The use of multimodal imaging has led to significant improvements in the diagnosis and treatment of many diseases. Similar to clinical practice, some works have demonstrated the benefits of multimodal fusion for automatic segmentation and classification using deep learning-based methods. However, current segmentation methods are limited to fusion of modalities with the same dimensionality (e.g., 3D+3D, 2D+2D), which is not always possible, and the fusion strategies implemented by classification methods are incompatible with localization tasks. In this work, we propose a novel deep learning-based framework for the fusion of multimodal data with heterogeneous dimensionality (e.g., 3D+2D) that is compatible with localization tasks. The proposed framework extracts the features of the different modalities and projects them into the common feature subspace. The projected features are then fused and further processed to obtain the final prediction. The framework was validated on the following tasks: segmentation of geographic atrophy (GA), a late-stage manifestation of age-related macular degeneration, and segmentation of retinal blood vessels (RBV) in multimodal retinal imaging. Our results show that the proposed method outperforms the state-of-the-art monomodal methods on GA and RBV segmentation by up to 3.10% and 4.64% Dice, respectively.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
SAMedOCT: Adapting Segment Anything Model (SAM) for Retinal OCT
Aug 31, 2023Abstract:The Segment Anything Model (SAM) has gained significant attention in the field of image segmentation due to its impressive capabilities and prompt-based interface. While SAM has already been extensively evaluated in various domains, its adaptation to retinal OCT scans remains unexplored. To bridge this research gap, we conduct a comprehensive evaluation of SAM and its adaptations on a large-scale public dataset of OCTs from RETOUCH challenge. Our evaluation covers diverse retinal diseases, fluid compartments, and device vendors, comparing SAM against state-of-the-art retinal fluid segmentation methods. Through our analysis, we showcase adapted SAM's efficacy as a powerful segmentation model in retinal OCT scans, although still lagging behind established methods in some circumstances. The findings highlight SAM's adaptability and robustness, showcasing its utility as a valuable tool in retinal OCT image analysis and paving the way for further advancements in this domain.
Self-supervised learning via inter-modal reconstruction and feature projection networks for label-efficient 3D-to-2D segmentation
Jul 13, 2023Abstract:Deep learning has become a valuable tool for the automation of certain medical image segmentation tasks, significantly relieving the workload of medical specialists. Some of these tasks require segmentation to be performed on a subset of the input dimensions, the most common case being 3D-to-2D. However, the performance of existing methods is strongly conditioned by the amount of labeled data available, as there is currently no data efficient method, e.g. transfer learning, that has been validated on these tasks. In this work, we propose a novel convolutional neural network (CNN) and self-supervised learning (SSL) method for label-efficient 3D-to-2D segmentation. The CNN is composed of a 3D encoder and a 2D decoder connected by novel 3D-to-2D blocks. The SSL method consists of reconstructing image pairs of modalities with different dimensionality. The approach has been validated in two tasks with clinical relevance: the en-face segmentation of geographic atrophy and reticular pseudodrusen in optical coherence tomography. Results on different datasets demonstrate that the proposed CNN significantly improves the state of the art in scenarios with limited labeled data by up to 8% in Dice score. Moreover, the proposed SSL method allows further improvement of this performance by up to 23%, and we show that the SSL is beneficial regardless of the network architecture.
AIROGS: Artificial Intelligence for RObust Glaucoma Screening Challenge
Feb 10, 2023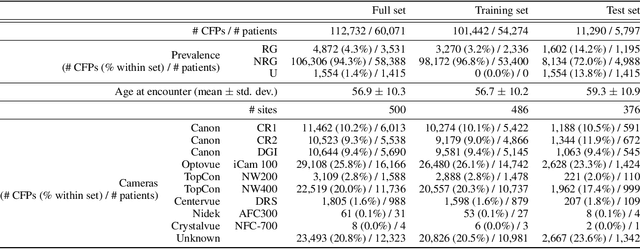
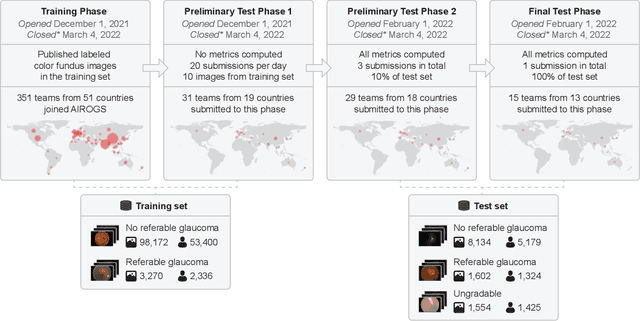
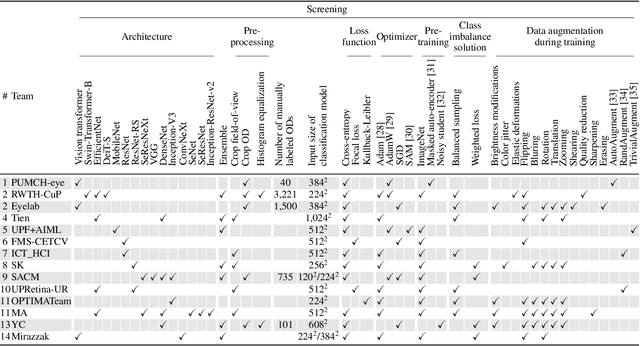
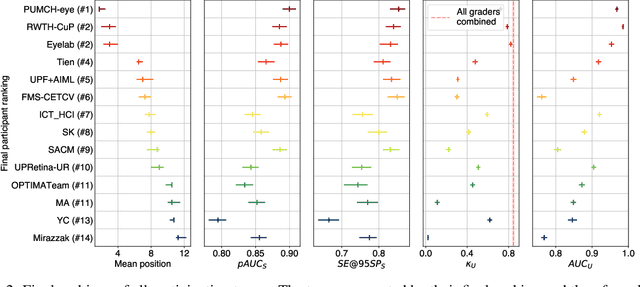
Abstract:The early detection of glaucoma is essential in preventing visual impairment. Artificial intelligence (AI) can be used to analyze color fundus photographs (CFPs) in a cost-effective manner, making glaucoma screening more accessible. While AI models for glaucoma screening from CFPs have shown promising results in laboratory settings, their performance decreases significantly in real-world scenarios due to the presence of out-of-distribution and low-quality images. To address this issue, we propose the Artificial Intelligence for Robust Glaucoma Screening (AIROGS) challenge. This challenge includes a large dataset of around 113,000 images from about 60,000 patients and 500 different screening centers, and encourages the development of algorithms that are robust to ungradable and unexpected input data. We evaluated solutions from 14 teams in this paper, and found that the best teams performed similarly to a set of 20 expert ophthalmologists and optometrists. The highest-scoring team achieved an area under the receiver operating characteristic curve of 0.99 (95% CI: 0.98-0.99) for detecting ungradable images on-the-fly. Additionally, many of the algorithms showed robust performance when tested on three other publicly available datasets. These results demonstrate the feasibility of robust AI-enabled glaucoma screening.
Segmentation of Bruch's Membrane in retinal OCT with AMD using anatomical priors and uncertainty quantification
Oct 30, 2022Abstract:Bruch's membrane (BM) segmentation on optical coherence tomography (OCT) is a pivotal step for the diagnosis and follow-up of age-related macular degeneration (AMD), one of the leading causes of blindness in the developed world. Automated BM segmentation methods exist, but they usually do not account for the anatomical coherence of the results, neither provide feedback on the confidence of the prediction. These factors limit the applicability of these systems in real-world scenarios. With this in mind, we propose an end-to-end deep learning method for automated BM segmentation in AMD patients. An Attention U-Net is trained to output a probability density function of the BM position, while taking into account the natural curvature of the surface. Besides the surface position, the method also estimates an A-scan wise uncertainty measure of the segmentation output. Subsequently, the A-scans with high uncertainty are interpolated using thin plate splines (TPS). We tested our method with ablation studies on an internal dataset with 138 patients covering all three AMD stages, and achieved a mean absolute localization error of 4.10 um. In addition, the proposed segmentation method was compared against the state-of-the-art methods and showed a superior performance on an external publicly available dataset from a different patient cohort and OCT device, demonstrating strong generalization ability.
SD-LayerNet: Semi-supervised retinal layer segmentation in OCT using disentangled representation with anatomical priors
Jul 01, 2022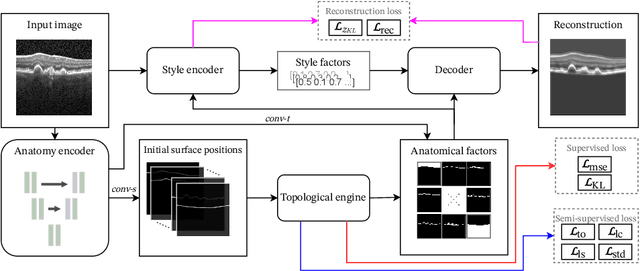
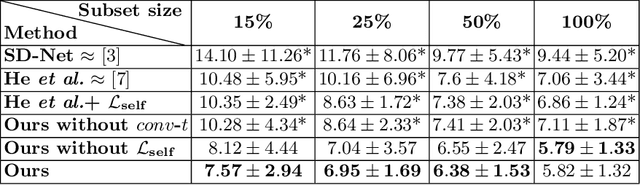
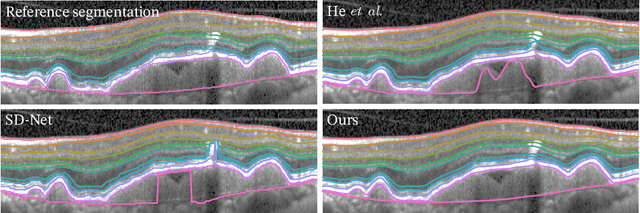
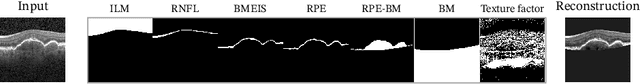
Abstract:Optical coherence tomography (OCT) is a non-invasive 3D modality widely used in ophthalmology for imaging the retina. Achieving automated, anatomically coherent retinal layer segmentation on OCT is important for the detection and monitoring of different retinal diseases, like Age-related Macular Disease (AMD) or Diabetic Retinopathy. However, the majority of state-of-the-art layer segmentation methods are based on purely supervised deep-learning, requiring a large amount of pixel-level annotated data that is expensive and hard to obtain. With this in mind, we introduce a semi-supervised paradigm into the retinal layer segmentation task that makes use of the information present in large-scale unlabeled datasets as well as anatomical priors. In particular, a novel fully differentiable approach is used for converting surface position regression into a pixel-wise structured segmentation, allowing to use both 1D surface and 2D layer representations in a coupled fashion to train the model. In particular, these 2D segmentations are used as anatomical factors that, together with learned style factors, compose disentangled representations used for reconstructing the input image. In parallel, we propose a set of anatomical priors to improve network training when a limited amount of labeled data is available. We demonstrate on the real-world dataset of scans with intermediate and wet-AMD that our method outperforms state-of-the-art when using our full training set, but more importantly largely exceeds state-of-the-art when it is trained with a fraction of the labeled data.
Deep Dirichlet uncertainty for unsupervised out-of-distribution detection of eye fundus photographs in glaucoma screening
Mar 18, 2022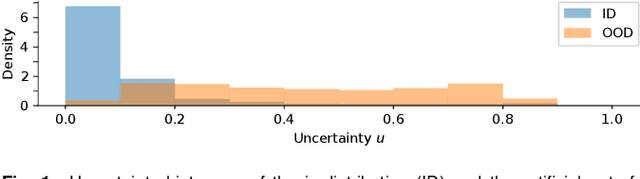
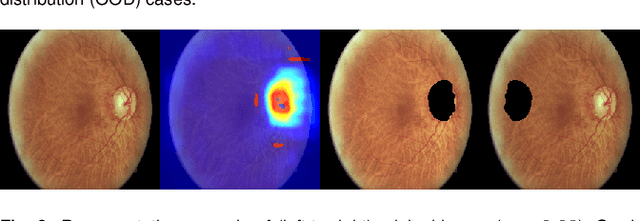
Abstract:The development of automatic tools for early glaucoma diagnosis with color fundus photographs can significantly reduce the impact of this disease. However, current state-of-the-art solutions are not robust to real-world scenarios, providing over-confident predictions for out-of-distribution cases. With this in mind, we propose a model based on the Dirichlet distribution that allows to obtain class-wise probabilities together with an uncertainty estimation without exposure to out-of-distribution cases. We demonstrate our approach on the AIROGS challenge. At the start of the final test phase (8 Feb. 2022), our method had the highest average score among all submissions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge