Christoph Grechenig
Deep Multimodal Fusion of Data with Heterogeneous Dimensionality via Projective Networks
Feb 02, 2024



Abstract:The use of multimodal imaging has led to significant improvements in the diagnosis and treatment of many diseases. Similar to clinical practice, some works have demonstrated the benefits of multimodal fusion for automatic segmentation and classification using deep learning-based methods. However, current segmentation methods are limited to fusion of modalities with the same dimensionality (e.g., 3D+3D, 2D+2D), which is not always possible, and the fusion strategies implemented by classification methods are incompatible with localization tasks. In this work, we propose a novel deep learning-based framework for the fusion of multimodal data with heterogeneous dimensionality (e.g., 3D+2D) that is compatible with localization tasks. The proposed framework extracts the features of the different modalities and projects them into the common feature subspace. The projected features are then fused and further processed to obtain the final prediction. The framework was validated on the following tasks: segmentation of geographic atrophy (GA), a late-stage manifestation of age-related macular degeneration, and segmentation of retinal blood vessels (RBV) in multimodal retinal imaging. Our results show that the proposed method outperforms the state-of-the-art monomodal methods on GA and RBV segmentation by up to 3.10% and 4.64% Dice, respectively.
Learning Spatio-Temporal Model of Disease Progression with NeuralODEs from Longitudinal Volumetric Data
Nov 08, 2022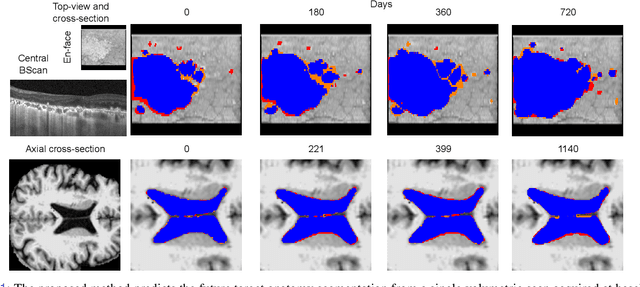
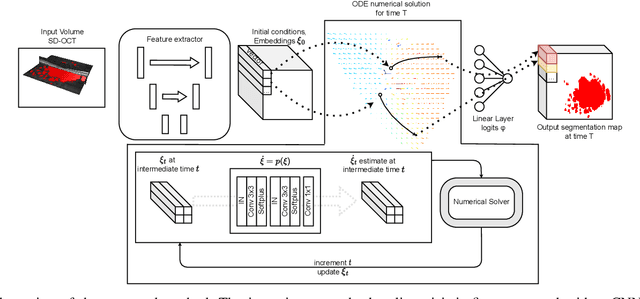
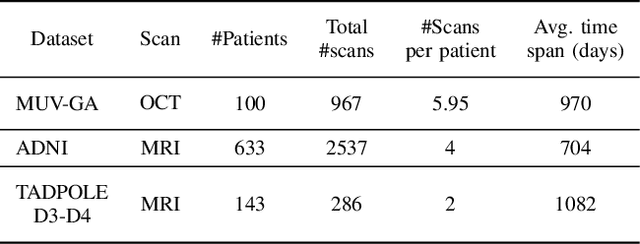
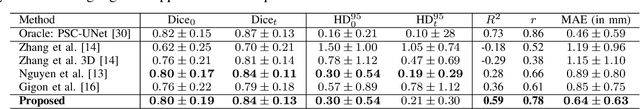
Abstract:Robust forecasting of the future anatomical changes inflicted by an ongoing disease is an extremely challenging task that is out of grasp even for experienced healthcare professionals. Such a capability, however, is of great importance since it can improve patient management by providing information on the speed of disease progression already at the admission stage, or it can enrich the clinical trials with fast progressors and avoid the need for control arms by the means of digital twins. In this work, we develop a deep learning method that models the evolution of age-related disease by processing a single medical scan and providing a segmentation of the target anatomy at a requested future point in time. Our method represents a time-invariant physical process and solves a large-scale problem of modeling temporal pixel-level changes utilizing NeuralODEs. In addition, we demonstrate the approaches to incorporate the prior domain-specific constraints into our method and define temporal Dice loss for learning temporal objectives. To evaluate the applicability of our approach across different age-related diseases and imaging modalities, we developed and tested the proposed method on the datasets with 967 retinal OCT volumes of 100 patients with Geographic Atrophy, and 2823 brain MRI volumes of 633 patients with Alzheimer's Disease. For Geographic Atrophy, the proposed method outperformed the related baseline models in the atrophy growth prediction. For Alzheimer's Disease, the proposed method demonstrated remarkable performance in predicting the brain ventricle changes induced by the disease, achieving the state-of-the-art result on TADPOLE challenge.
Modeling Disease Progression In Retinal OCTs With Longitudinal Self-Supervised Learning
Oct 24, 2019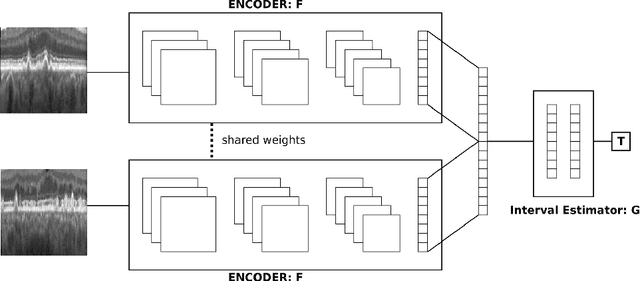
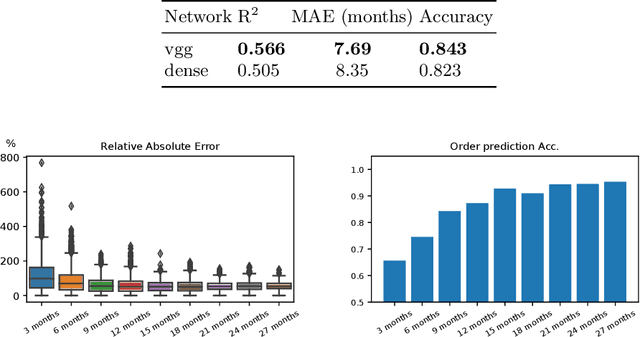
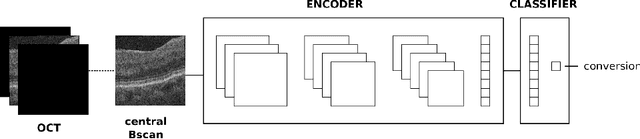
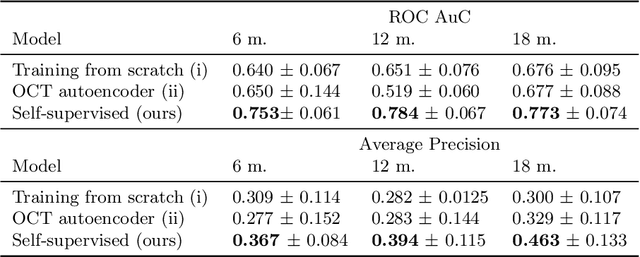
Abstract:Longitudinal imaging is capable of capturing the static ana\-to\-mi\-cal structures and the dynamic changes of the morphology resulting from aging or disease progression. Self-supervised learning allows to learn new representation from available large unlabelled data without any expert knowledge. We propose a deep learning self-supervised approach to model disease progression from longitudinal retinal optical coherence tomography (OCT). Our self-supervised model takes benefit from a generic time-related task, by learning to estimate the time interval between pairs of scans acquired from the same patient. This task is (i) easy to implement, (ii) allows to use irregularly sampled data, (iii) is tolerant to poor registration, and (iv) does not rely on additional annotations. This novel method learns a representation that focuses on progression specific information only, which can be transferred to other types of longitudinal problems. We transfer the learnt representation to a clinically highly relevant task of predicting the onset of an advanced stage of age-related macular degeneration within a given time interval based on a single OCT scan. The boost in prediction accuracy, in comparison to a network learned from scratch or transferred from traditional tasks, demonstrates that our pretrained self-supervised representation learns a clinically meaningful information.
U2-Net: A Bayesian U-Net model with epistemic uncertainty feedback for photoreceptor layer segmentation in pathological OCT scans
Jan 23, 2019



Abstract:In this paper, we introduce a Bayesian deep learning based model for segmenting the photoreceptor layer in pathological OCT scans. Our architecture provides accurate segmentations of the photoreceptor layer and produces pixel-wise epistemic uncertainty maps that highlight potential areas of pathologies or segmentation errors. We empirically evaluated this approach in two sets of pathological OCT scans of patients with age-related macular degeneration, retinal vein oclussion and diabetic macular edema, improving the performance of the baseline U-Net both in terms of the Dice index and the area under the precision/recall curve. We also observed that the uncertainty estimates were inversely correlated with the model performance, underlying its utility for highlighting areas where manual inspection/correction might be needed.
* Accepted for publication at IEEE International Symposium on Biomedical Imaging (ISBI) 2019
On orthogonal projections for dimension reduction and applications in variational loss functions for learning problems
Jan 22, 2019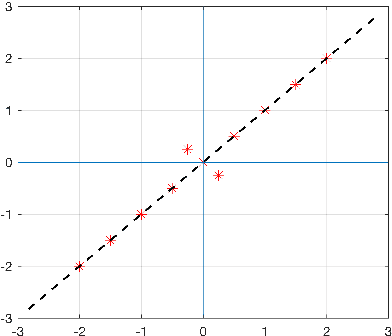
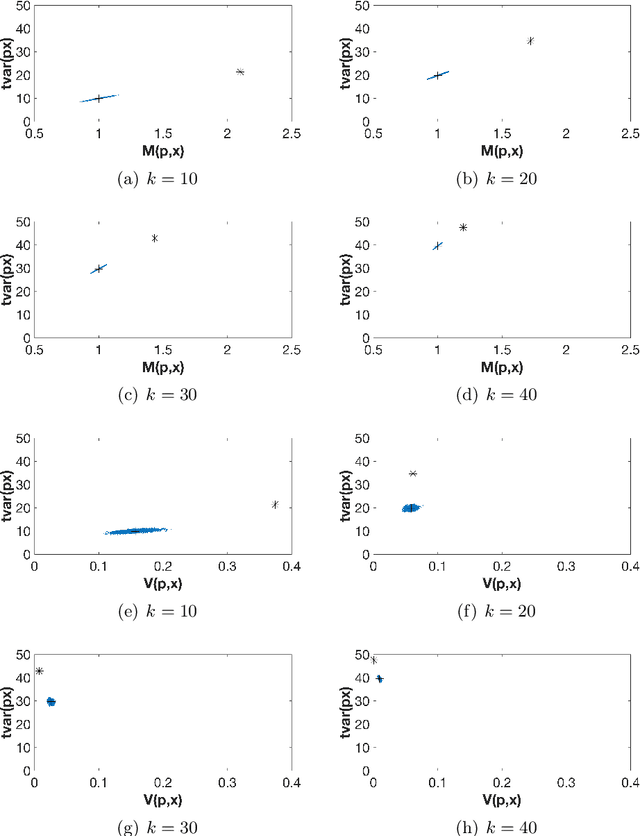
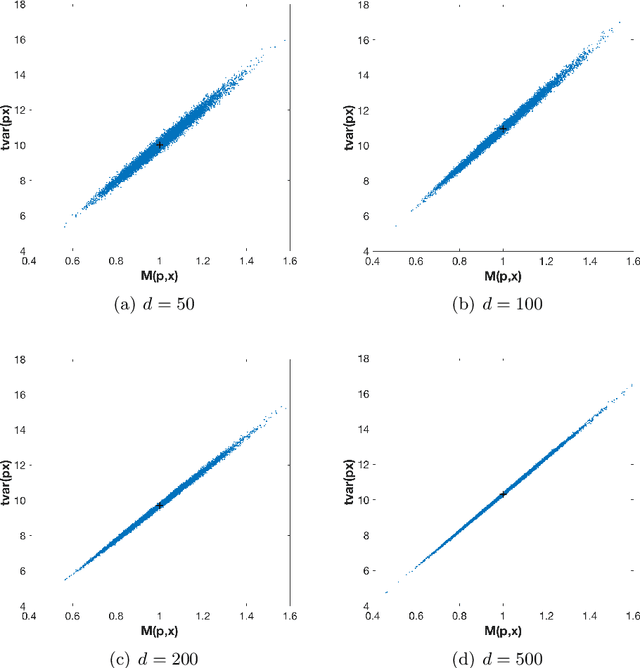
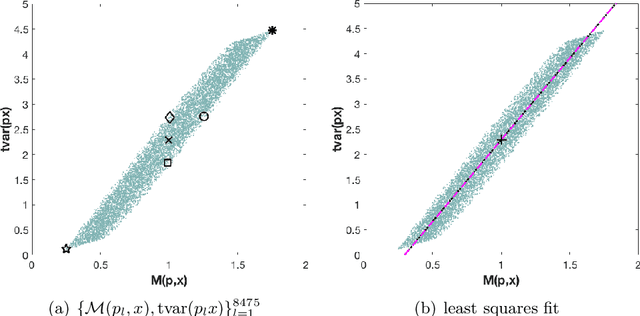
Abstract:The use of orthogonal projections on high-dimensional input and target data in learning frameworks is studied. First, we investigate the relations between two standard objectives in dimension reduction, maximizing variance and preservation of pairwise relative distances. The derivation of their asymptotic correlation and numerical experiments tell that a projection usually cannot satisfy both objectives. In a standard classification problem we determine projections on the input data that balance them and compare subsequent results. Next, we extend our application of orthogonal projections to deep learning frameworks. We introduce new variational loss functions that enable integration of additional information via transformations and projections of the target data. In two supervised learning problems, clinical image segmentation and music information classification, the application of the proposed loss functions increase the accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge