Sebastian Waldstein
U-Net with spatial pyramid pooling for drusen segmentation in optical coherence tomography
Dec 11, 2019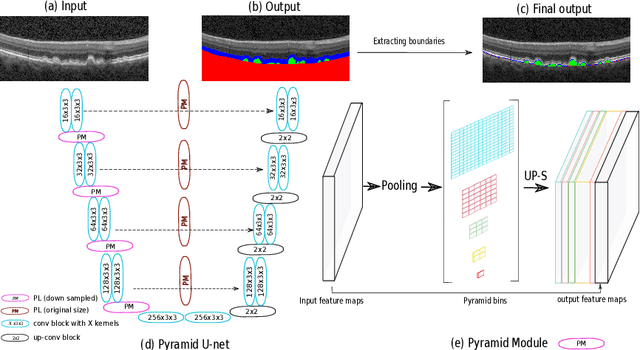

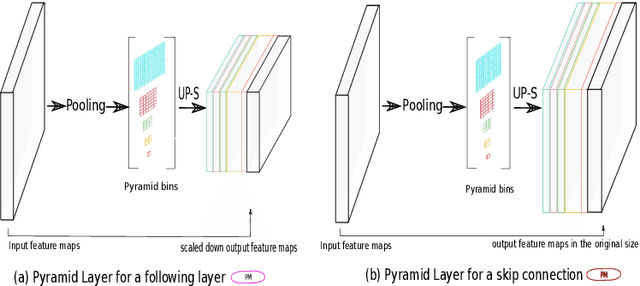
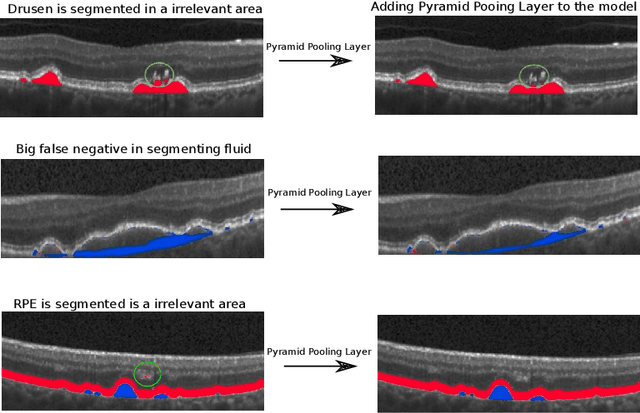
Abstract:The presence of drusen is the main hallmark of early/intermediate age-related macular degeneration (AMD). Therefore, automated drusen segmentation is an important step in image-guided management of AMD. There are two common approaches to drusen segmentation. In the first, the drusen are segmented directly as a binary classification task. In the second approach, the surrounding retinal layers (outer boundary retinal pigment epithelium (OBRPE) and Bruch's membrane (BM)) are segmented and the remaining space between these two layers is extracted as drusen. In this work, we extend the standard U-Net architecture with spatial pyramid pooling components to introduce global feature context. We apply the model to the task of segmenting drusen together with BM and OBRPE. The proposed network was trained and evaluated on a longitudinal OCT dataset of 425 scans from 38 patients with early/intermediate AMD. This preliminary study showed that the proposed network consistently outperformed the standard U-net model.
Multiclass segmentation as multitask learning for drusen segmentation in retinal optical coherence tomography
Jul 24, 2019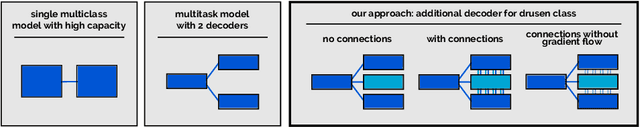
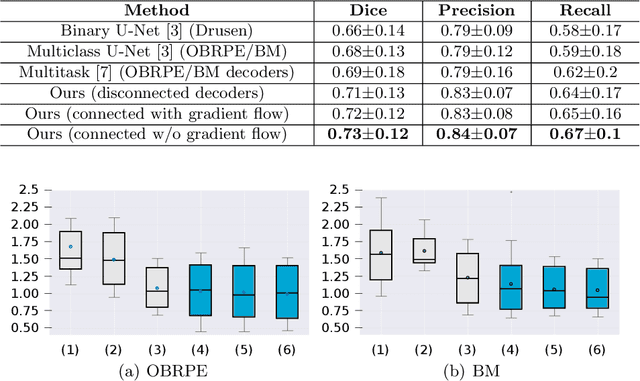
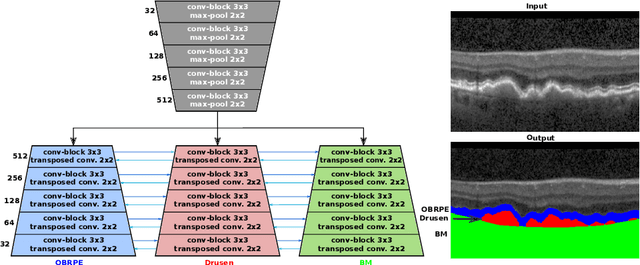
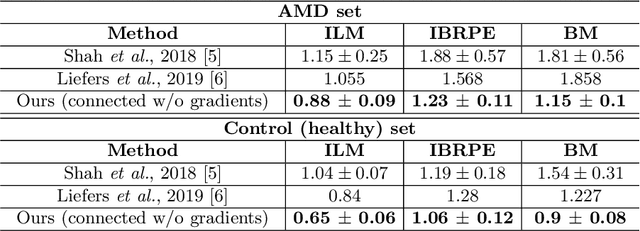
Abstract:Automated drusen segmentation in retinal optical coherence tomography (OCT) scans is relevant for understanding age-related macular degeneration (AMD) risk and progression. This task is usually performed by segmenting the top/bottom anatomical interfaces that define drusen, the outer boundary of the retinal pigment epithelium (OBRPE) and the Bruch's membrane (BM), respectively. In this paper we propose a novel multi-decoder architecture that tackles drusen segmentation as a multitask problem. Instead of training a multiclass model for OBRPE/BM segmentation, we use one decoder per target class and an extra one aiming for the area between the layers. We also introduce connections between each class-specific branch and the additional decoder to increase the regularization effect of this surrogate task. We validated our approach on private/public data sets with 166 early/intermediate AMD Spectralis, and 200 AMD and control Bioptigen OCT volumes, respectively. Our method consistently outperformed several baselines in both layer and drusen segmentation evaluations.
Using CycleGANs for effectively reducing image variability across OCT devices and improving retinal fluid segmentation
Jan 25, 2019


Abstract:Optical coherence tomography (OCT) has become the most important imaging modality in ophthalmology. A substantial amount of research has recently been devoted to the development of machine learning (ML) models for the identification and quantification of pathological features in OCT images. Among the several sources of variability the ML models have to deal with, a major factor is the acquisition device, which can limit the ML model's generalizability. In this paper, we propose to reduce the image variability across different OCT devices (Spectralis and Cirrus) by using CycleGAN, an unsupervised unpaired image transformation algorithm. The usefulness of this approach is evaluated in the setting of retinal fluid segmentation, namely intraretinal cystoid fluid (IRC) and subretinal fluid (SRF). First, we train a segmentation model on images acquired with a source OCT device. Then we evaluate the model on (1) source, (2) target and (3) transformed versions of the target OCT images. The presented transformation strategy shows an F1 score of 0.4 (0.51) for IRC (SRF) segmentations. Compared with traditional transformation approaches, this means an F1 score gain of 0.2 (0.12).
U2-Net: A Bayesian U-Net model with epistemic uncertainty feedback for photoreceptor layer segmentation in pathological OCT scans
Jan 23, 2019



Abstract:In this paper, we introduce a Bayesian deep learning based model for segmenting the photoreceptor layer in pathological OCT scans. Our architecture provides accurate segmentations of the photoreceptor layer and produces pixel-wise epistemic uncertainty maps that highlight potential areas of pathologies or segmentation errors. We empirically evaluated this approach in two sets of pathological OCT scans of patients with age-related macular degeneration, retinal vein oclussion and diabetic macular edema, improving the performance of the baseline U-Net both in terms of the Dice index and the area under the precision/recall curve. We also observed that the uncertainty estimates were inversely correlated with the model performance, underlying its utility for highlighting areas where manual inspection/correction might be needed.
* Accepted for publication at IEEE International Symposium on Biomedical Imaging (ISBI) 2019
Identifying and Categorizing Anomalies in Retinal Imaging Data
Dec 02, 2016
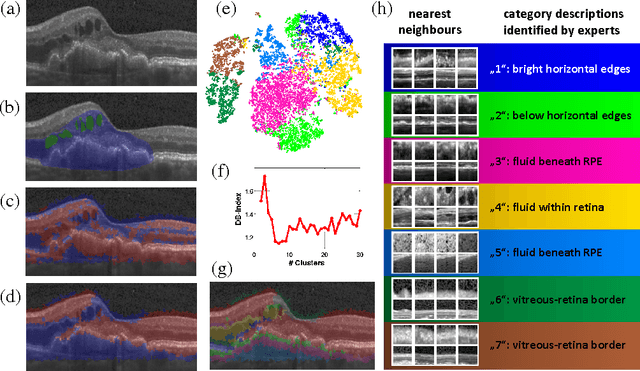
Abstract:The identification and quantification of markers in medical images is critical for diagnosis, prognosis and management of patients in clinical practice. Supervised- or weakly supervised training enables the detection of findings that are known a priori. It does not scale well, and a priori definition limits the vocabulary of markers to known entities reducing the accuracy of diagnosis and prognosis. Here, we propose the identification of anomalies in large-scale medical imaging data using healthy examples as a reference. We detect and categorize candidates for anomaly findings untypical for the observed data. A deep convolutional autoencoder is trained on healthy retinal images. The learned model generates a new feature representation, and the distribution of healthy retinal patches is estimated by a one-class support vector machine. Results demonstrate that we can identify pathologic regions in images without using expert annotations. A subsequent clustering categorizes findings into clinically meaningful classes. In addition the learned features outperform standard embedding approaches in a classification task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge