Thomas Schlegl
Department of Biomedical Imaging and Image-guided Therapy, Computational Imaging Research Lab, Medical University Vienna, Austria, Christian Doppler Laboratory for Ophthalmic Image Analysis, Department of Ophthalmology and Optometry, Medical University Vienna, Austria
Data-centric AI approach to improve optic nerve head segmentation and localization in OCT en face images
Aug 08, 2022

Abstract:The automatic detection and localization of anatomical features in retinal imaging data are relevant for many aspects. In this work, we follow a data-centric approach to optimize classifier training for optic nerve head detection and localization in optical coherence tomography en face images of the retina. We examine the effect of domain knowledge driven spatial complexity reduction on the resulting optic nerve head segmentation and localization performance. We present a machine learning approach for segmenting optic nerve head in 2D en face projections of 3D widefield swept source optical coherence tomography scans that enables the automated assessment of large amounts of data. Evaluation on manually annotated 2D en face images of the retina demonstrates that training of a standard U-Net can yield improved optic nerve head segmentation and localization performance when the underlying pixel-level binary classification task is spatially relaxed through domain knowledge.
Exploiting Epistemic Uncertainty of Anatomy Segmentation for Anomaly Detection in Retinal OCT
May 29, 2019



Abstract:Diagnosis and treatment guidance are aided by detecting relevant biomarkers in medical images. Although supervised deep learning can perform accurate segmentation of pathological areas, it is limited by requiring a-priori definitions of these regions, large-scale annotations, and a representative patient cohort in the training set. In contrast, anomaly detection is not limited to specific definitions of pathologies and allows for training on healthy samples without annotation. Anomalous regions can then serve as candidates for biomarker discovery. Knowledge about normal anatomical structure brings implicit information for detecting anomalies. We propose to take advantage of this property using bayesian deep learning, based on the assumption that epistemic uncertainties will correlate with anatomical deviations from a normal training set. A Bayesian U-Net is trained on a well-defined healthy environment using weak labels of healthy anatomy produced by existing methods. At test time, we capture epistemic uncertainty estimates of our model using Monte Carlo dropout. A novel post-processing technique is then applied to exploit these estimates and transfer their layered appearance to smooth blob-shaped segmentations of the anomalies. We experimentally validated this approach in retinal optical coherence tomography (OCT) images, using weak labels of retinal layers. Our method achieved a Dice index of 0.789 in an independent anomaly test set of age-related macular degeneration (AMD) cases. The resulting segmentations allowed very high accuracy for separating healthy and diseased cases with late wet AMD, dry geographic atrophy (GA), diabetic macular edema (DME) and retinal vein occlusion (RVO). Finally, we qualitatively observed that our approach can also detect other deviations in normal scans such as cut edge artifacts.
Unsupervised Identification of Disease Marker Candidates in Retinal OCT Imaging Data
Oct 31, 2018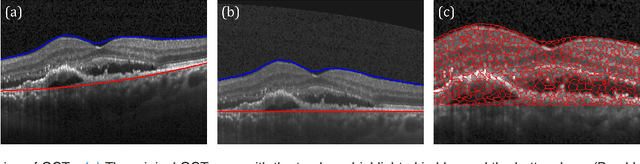
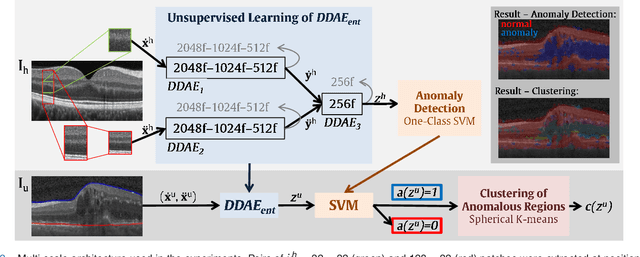
Abstract:The identification and quantification of markers in medical images is critical for diagnosis, prognosis, and disease management. Supervised machine learning enables the detection and exploitation of findings that are known a priori after annotation of training examples by experts. However, supervision does not scale well, due to the amount of necessary training examples, and the limitation of the marker vocabulary to known entities. In this proof-of-concept study, we propose unsupervised identification of anomalies as candidates for markers in retinal Optical Coherence Tomography (OCT) imaging data without a constraint to a priori definitions. We identify and categorize marker candidates occurring frequently in the data, and demonstrate that these markers show predictive value in the task of detecting disease. A careful qualitative analysis of the identified data driven markers reveals how their quantifiable occurrence aligns with our current understanding of disease course, in early- and late age-related macular degeneration (AMD) patients. A multi-scale deep denoising autoencoder is trained on healthy images, and a one-class support vector machine identifies anomalies in new data. Clustering in the anomalies identifies stable categories. Using these markers to classify healthy-, early AMD- and late AMD cases yields an accuracy of 81.40%. In a second binary classification experiment on a publicly available data set (healthy vs. intermediate AMD) the model achieves an area under the ROC curve of 0.944.
Fully Automated Segmentation of Hyperreflective Foci in Optical Coherence Tomography Images
May 08, 2018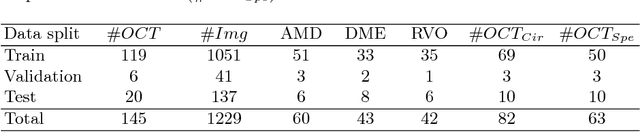
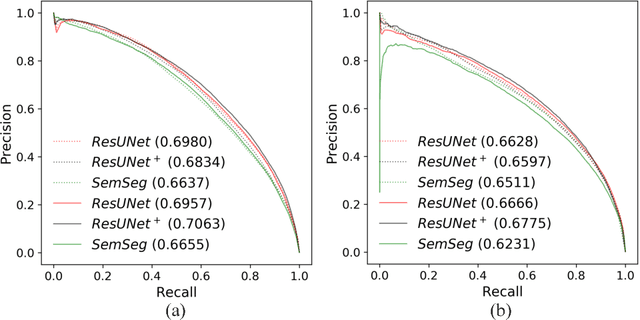
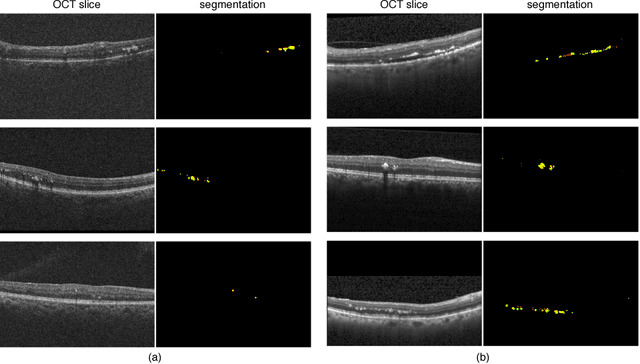
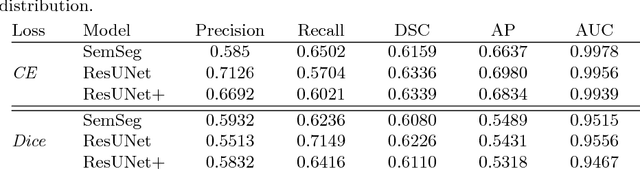
Abstract:The automatic detection of disease related entities in retinal imaging data is relevant for disease- and treatment monitoring. It enables the quantitative assessment of large amounts of data and the corresponding study of disease characteristics. The presence of hyperreflective foci (HRF) is related to disease progression in various retinal diseases. Manual identification of HRF in spectral-domain optical coherence tomography (SD-OCT) scans is error-prone and tedious. We present a fully automated machine learning approach for segmenting HRF in SD-OCT scans. Evaluation on annotated OCT images of the retina demonstrates that a residual U-Net allows to segment HRF with high accuracy. As our dataset comprised data from different retinal diseases including age-related macular degeneration, diabetic macular edema and retinal vein occlusion, the algorithm can safely be applied in all of them though different pathophysiological origins are known.
Unsupervised Anomaly Detection with Generative Adversarial Networks to Guide Marker Discovery
Mar 17, 2017
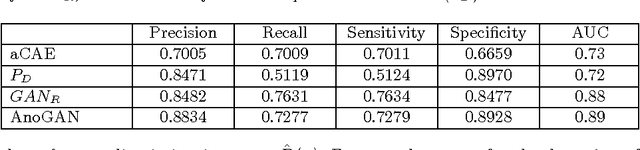
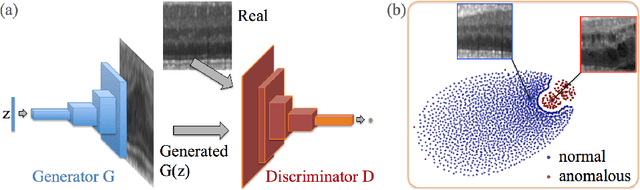

Abstract:Obtaining models that capture imaging markers relevant for disease progression and treatment monitoring is challenging. Models are typically based on large amounts of data with annotated examples of known markers aiming at automating detection. High annotation effort and the limitation to a vocabulary of known markers limit the power of such approaches. Here, we perform unsupervised learning to identify anomalies in imaging data as candidates for markers. We propose AnoGAN, a deep convolutional generative adversarial network to learn a manifold of normal anatomical variability, accompanying a novel anomaly scoring scheme based on the mapping from image space to a latent space. Applied to new data, the model labels anomalies, and scores image patches indicating their fit into the learned distribution. Results on optical coherence tomography images of the retina demonstrate that the approach correctly identifies anomalous images, such as images containing retinal fluid or hyperreflective foci.
Identifying and Categorizing Anomalies in Retinal Imaging Data
Dec 02, 2016
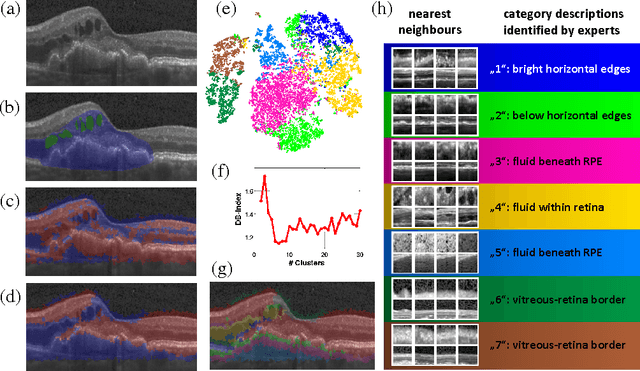
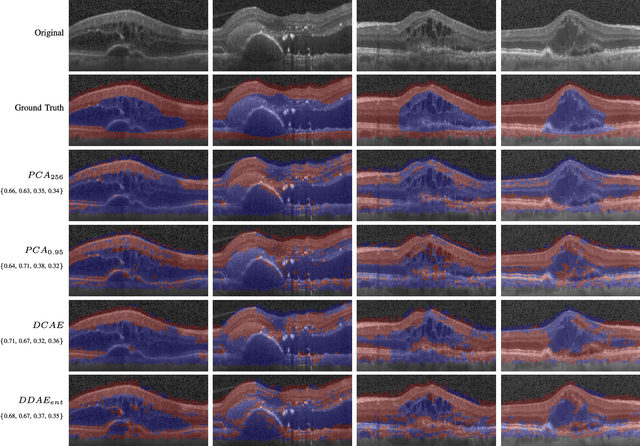
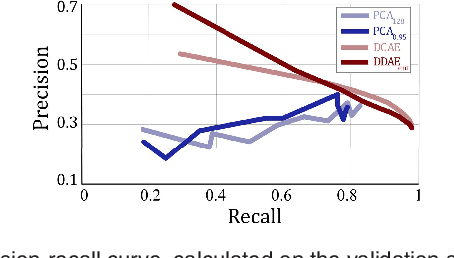
Abstract:The identification and quantification of markers in medical images is critical for diagnosis, prognosis and management of patients in clinical practice. Supervised- or weakly supervised training enables the detection of findings that are known a priori. It does not scale well, and a priori definition limits the vocabulary of markers to known entities reducing the accuracy of diagnosis and prognosis. Here, we propose the identification of anomalies in large-scale medical imaging data using healthy examples as a reference. We detect and categorize candidates for anomaly findings untypical for the observed data. A deep convolutional autoencoder is trained on healthy retinal images. The learned model generates a new feature representation, and the distribution of healthy retinal patches is estimated by a one-class support vector machine. Results demonstrate that we can identify pathologic regions in images without using expert annotations. A subsequent clustering categorizes findings into clinically meaningful classes. In addition the learned features outperform standard embedding approaches in a classification task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge