Hrvoje Bogunovic
*: shared first/last authors
Transformer-based end-to-end classification of variable-length volumetric data
Jul 21, 2023



Abstract:The automatic classification of 3D medical data is memory-intensive. Also, variations in the number of slices between samples is common. Na\"ive solutions such as subsampling can solve these problems, but at the cost of potentially eliminating relevant diagnosis information. Transformers have shown promising performance for sequential data analysis. However, their application for long sequences is data, computationally, and memory demanding. In this paper, we propose an end-to-end Transformer-based framework that allows to classify volumetric data of variable length in an efficient fashion. Particularly, by randomizing the input volume-wise resolution(#slices) during training, we enhance the capacity of the learnable positional embedding assigned to each volume slice. Consequently, the accumulated positional information in each positional embedding can be generalized to the neighbouring slices, even for high-resolution volumes at the test time. By doing so, the model will be more robust to variable volume length and amenable to different computational budgets. We evaluated the proposed approach in retinal OCT volume classification and achieved 21.96% average improvement in balanced accuracy on a 9-class diagnostic task, compared to state-of-the-art video transformers. Our findings show that varying the volume-wise resolution of the input during training results in more informative volume representation as compared to training with fixed number of slices per volume.
Learning Spatio-Temporal Model of Disease Progression with NeuralODEs from Longitudinal Volumetric Data
Nov 08, 2022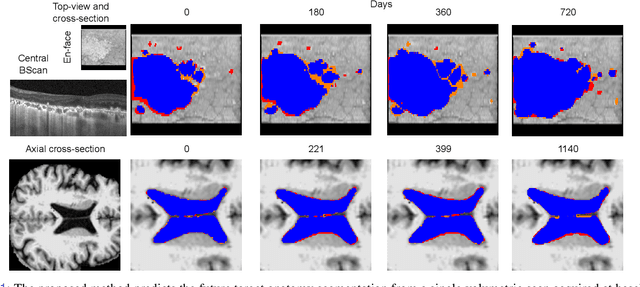
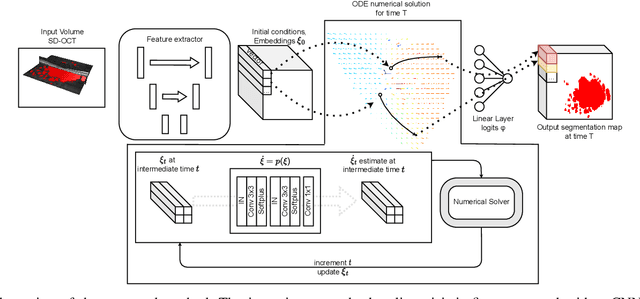
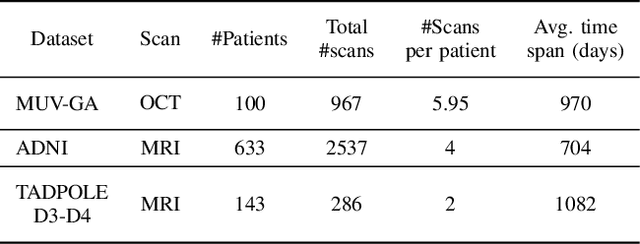
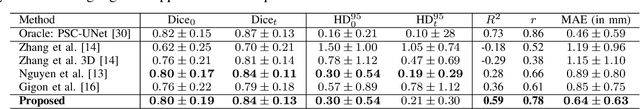
Abstract:Robust forecasting of the future anatomical changes inflicted by an ongoing disease is an extremely challenging task that is out of grasp even for experienced healthcare professionals. Such a capability, however, is of great importance since it can improve patient management by providing information on the speed of disease progression already at the admission stage, or it can enrich the clinical trials with fast progressors and avoid the need for control arms by the means of digital twins. In this work, we develop a deep learning method that models the evolution of age-related disease by processing a single medical scan and providing a segmentation of the target anatomy at a requested future point in time. Our method represents a time-invariant physical process and solves a large-scale problem of modeling temporal pixel-level changes utilizing NeuralODEs. In addition, we demonstrate the approaches to incorporate the prior domain-specific constraints into our method and define temporal Dice loss for learning temporal objectives. To evaluate the applicability of our approach across different age-related diseases and imaging modalities, we developed and tested the proposed method on the datasets with 967 retinal OCT volumes of 100 patients with Geographic Atrophy, and 2823 brain MRI volumes of 633 patients with Alzheimer's Disease. For Geographic Atrophy, the proposed method outperformed the related baseline models in the atrophy growth prediction. For Alzheimer's Disease, the proposed method demonstrated remarkable performance in predicting the brain ventricle changes induced by the disease, achieving the state-of-the-art result on TADPOLE challenge.
Segmentation of Bruch's Membrane in retinal OCT with AMD using anatomical priors and uncertainty quantification
Oct 30, 2022Abstract:Bruch's membrane (BM) segmentation on optical coherence tomography (OCT) is a pivotal step for the diagnosis and follow-up of age-related macular degeneration (AMD), one of the leading causes of blindness in the developed world. Automated BM segmentation methods exist, but they usually do not account for the anatomical coherence of the results, neither provide feedback on the confidence of the prediction. These factors limit the applicability of these systems in real-world scenarios. With this in mind, we propose an end-to-end deep learning method for automated BM segmentation in AMD patients. An Attention U-Net is trained to output a probability density function of the BM position, while taking into account the natural curvature of the surface. Besides the surface position, the method also estimates an A-scan wise uncertainty measure of the segmentation output. Subsequently, the A-scans with high uncertainty are interpolated using thin plate splines (TPS). We tested our method with ablation studies on an internal dataset with 138 patients covering all three AMD stages, and achieved a mean absolute localization error of 4.10 um. In addition, the proposed segmentation method was compared against the state-of-the-art methods and showed a superior performance on an external publicly available dataset from a different patient cohort and OCT device, demonstrating strong generalization ability.
SD-LayerNet: Semi-supervised retinal layer segmentation in OCT using disentangled representation with anatomical priors
Jul 01, 2022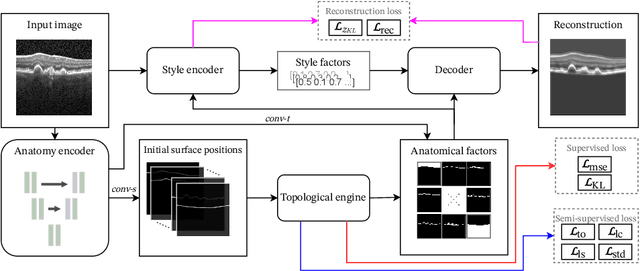
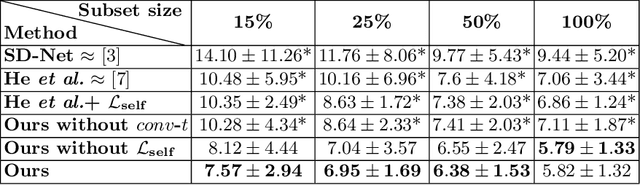
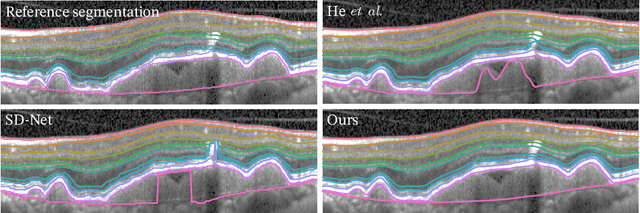
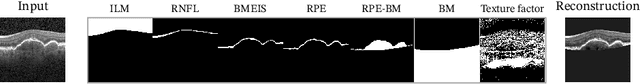
Abstract:Optical coherence tomography (OCT) is a non-invasive 3D modality widely used in ophthalmology for imaging the retina. Achieving automated, anatomically coherent retinal layer segmentation on OCT is important for the detection and monitoring of different retinal diseases, like Age-related Macular Disease (AMD) or Diabetic Retinopathy. However, the majority of state-of-the-art layer segmentation methods are based on purely supervised deep-learning, requiring a large amount of pixel-level annotated data that is expensive and hard to obtain. With this in mind, we introduce a semi-supervised paradigm into the retinal layer segmentation task that makes use of the information present in large-scale unlabeled datasets as well as anatomical priors. In particular, a novel fully differentiable approach is used for converting surface position regression into a pixel-wise structured segmentation, allowing to use both 1D surface and 2D layer representations in a coupled fashion to train the model. In particular, these 2D segmentations are used as anatomical factors that, together with learned style factors, compose disentangled representations used for reconstructing the input image. In parallel, we propose a set of anatomical priors to improve network training when a limited amount of labeled data is available. We demonstrate on the real-world dataset of scans with intermediate and wet-AMD that our method outperforms state-of-the-art when using our full training set, but more importantly largely exceeds state-of-the-art when it is trained with a fraction of the labeled data.
Deep Dirichlet uncertainty for unsupervised out-of-distribution detection of eye fundus photographs in glaucoma screening
Mar 18, 2022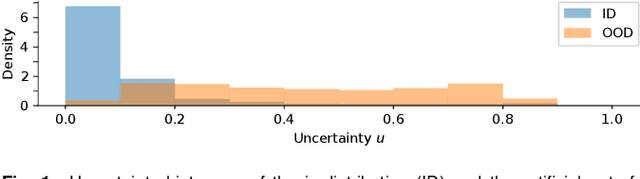
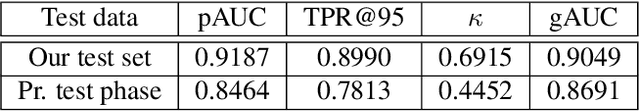
Abstract:The development of automatic tools for early glaucoma diagnosis with color fundus photographs can significantly reduce the impact of this disease. However, current state-of-the-art solutions are not robust to real-world scenarios, providing over-confident predictions for out-of-distribution cases. With this in mind, we propose a model based on the Dirichlet distribution that allows to obtain class-wise probabilities together with an uncertainty estimation without exposure to out-of-distribution cases. We demonstrate our approach on the AIROGS challenge. At the start of the final test phase (8 Feb. 2022), our method had the highest average score among all submissions.
GAMMA Challenge:Glaucoma grAding from Multi-Modality imAges
Feb 16, 2022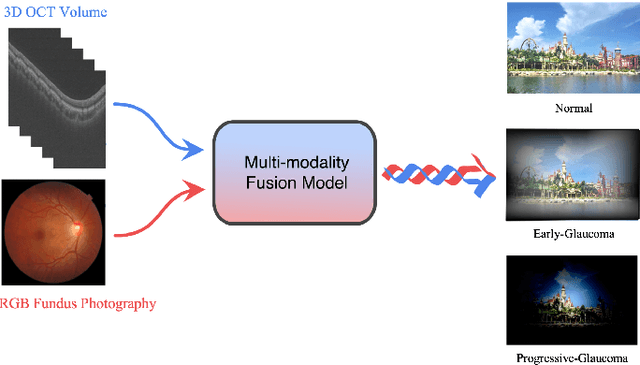
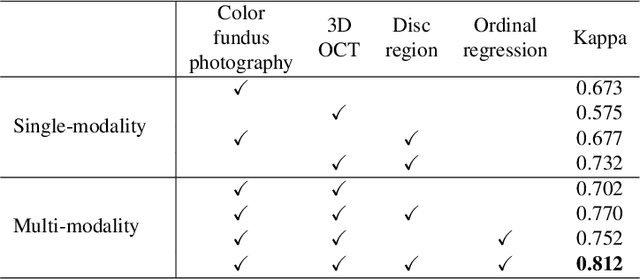
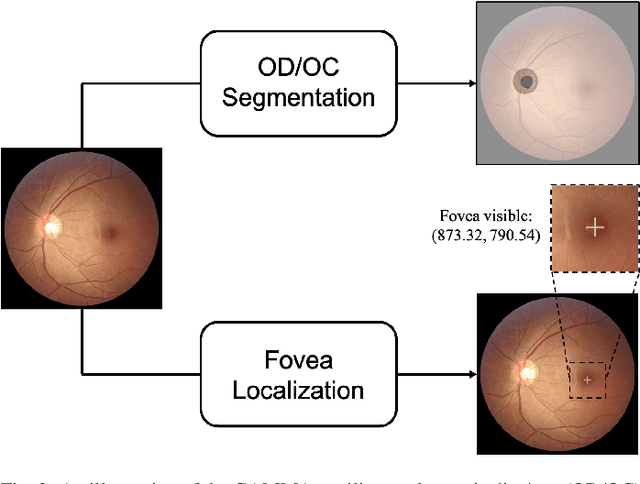
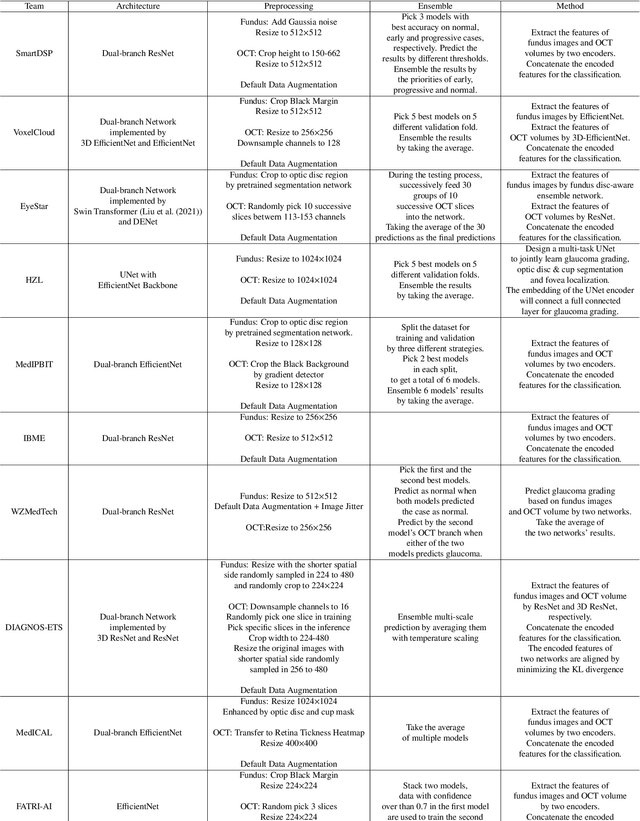
Abstract:Color fundus photography and Optical Coherence Tomography (OCT) are the two most cost-effective tools for glaucoma screening. Both two modalities of images have prominent biomarkers to indicate glaucoma suspected. Clinically, it is often recommended to take both of the screenings for a more accurate and reliable diagnosis. However, although numerous algorithms are proposed based on fundus images or OCT volumes in computer-aided diagnosis, there are still few methods leveraging both of the modalities for the glaucoma assessment. Inspired by the success of Retinal Fundus Glaucoma Challenge (REFUGE) we held previously, we set up the Glaucoma grAding from Multi-Modality imAges (GAMMA) Challenge to encourage the development of fundus \& OCT-based glaucoma grading. The primary task of the challenge is to grade glaucoma from both the 2D fundus images and 3D OCT scanning volumes. As part of GAMMA, we have publicly released a glaucoma annotated dataset with both 2D fundus color photography and 3D OCT volumes, which is the first multi-modality dataset for glaucoma grading. In addition, an evaluation framework is also established to evaluate the performance of the submitted methods. During the challenge, 1272 results were submitted, and finally, top-10 teams were selected to the final stage. We analysis their results and summarize their methods in the paper. Since all these teams submitted their source code in the challenge, a detailed ablation study is also conducted to verify the effectiveness of the particular modules proposed. We find many of the proposed techniques are practical for the clinical diagnosis of glaucoma. As the first in-depth study of fundus \& OCT multi-modality glaucoma grading, we believe the GAMMA Challenge will be an essential starting point for future research.
Projective Skip-Connections for Segmentation Along a Subset of Dimensions in Retinal OCT
Aug 02, 2021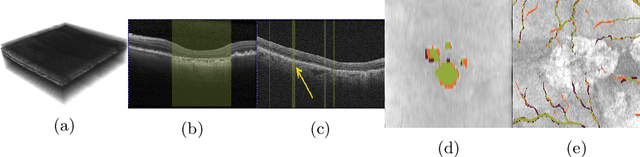
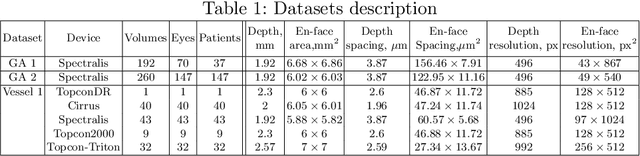
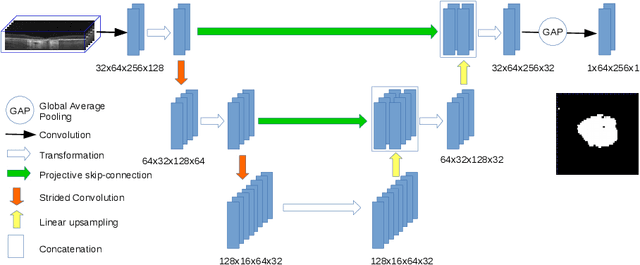
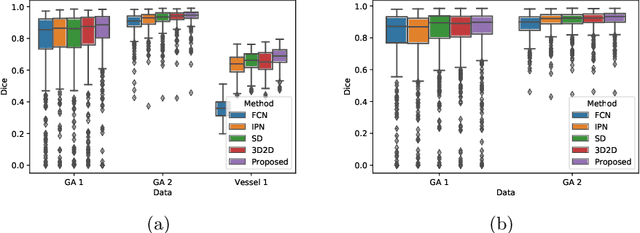
Abstract:In medical imaging, there are clinically relevant segmentation tasks where the output mask is a projection to a subset of input image dimensions. In this work, we propose a novel convolutional neural network architecture that can effectively learn to produce a lower-dimensional segmentation mask than the input image. The network restores encoded representation only in a subset of input spatial dimensions and keeps the representation unchanged in the others. The newly proposed projective skip-connections allow linking the encoder and decoder in a UNet-like structure. We evaluated the proposed method on two clinically relevant tasks in retinal Optical Coherence Tomography (OCT): geographic atrophy and retinal blood vessel segmentation. The proposed method outperformed the current state-of-the-art approaches on all the OCT datasets used, consisting of 3D volumes and corresponding 2D en-face masks. The proposed architecture fills the methodological gap between image classification and ND image segmentation.
AGE Challenge: Angle Closure Glaucoma Evaluation in Anterior Segment Optical Coherence Tomography
May 05, 2020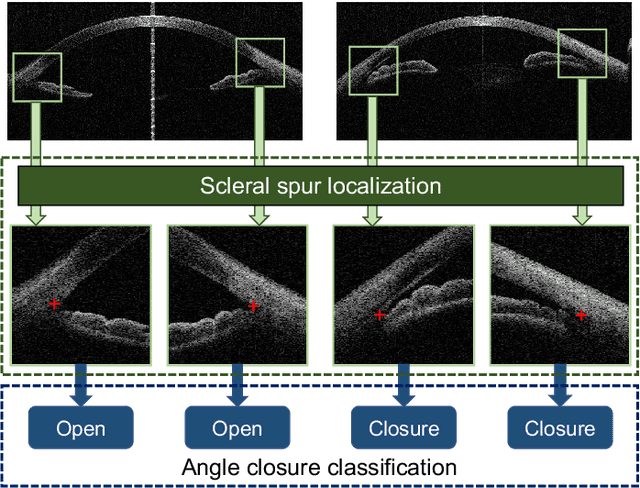
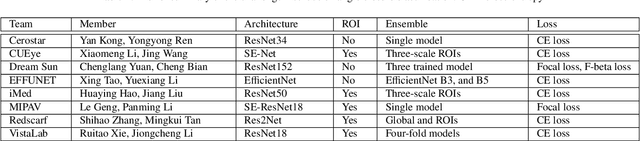
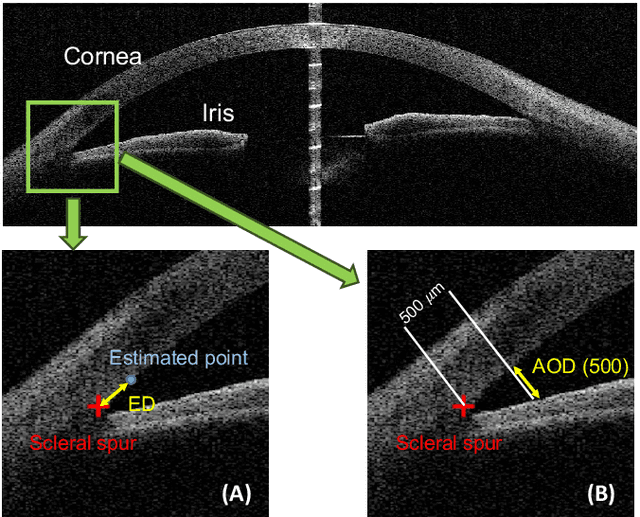
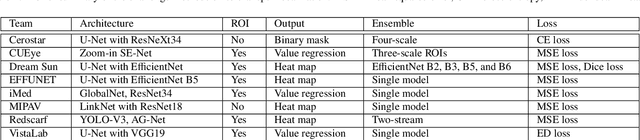
Abstract:Angle closure glaucoma (ACG) is a more aggressive disease than open-angle glaucoma, where the abnormal anatomical structures of the anterior chamber angle (ACA) may cause an elevated intraocular pressure and gradually leads to glaucomatous optic neuropathy and eventually to visual impairment and blindness. Anterior Segment Optical Coherence Tomography (AS-OCT) imaging provides a fast and contactless way to discriminate angle closure from open angle. Although many medical image analysis algorithms have been developed for glaucoma diagnosis, only a few studies have focused on AS-OCT imaging. In particular, there is no public AS-OCT dataset available for evaluating the existing methods in a uniform way, which limits the progress in the development of automated techniques for angle closure detection and assessment. To address this, we organized the Angle closure Glaucoma Evaluation challenge (AGE), held in conjunction with MICCAI 2019. The AGE challenge consisted of two tasks: scleral spur localization and angle closure classification. For this challenge, we released a large data of 4800 annotated AS-OCT images from 199 patients, and also proposed an evaluation framework to benchmark and compare different models. During the AGE challenge, over 200 teams registered online, and more than 1100 results were submitted for online evaluation. Finally, eight teams participated in the onsite challenge. In this paper, we summarize these eight onsite challenge methods and analyze their corresponding results in the two tasks. We further discuss limitations and future directions. In the AGE challenge, the top-performing approach had an average Euclidean Distance of 10 pixel in scleral spur localization, while in the task of angle closure classification, all the algorithms achieved the satisfactory performances, especially, 100% accuracy rate for top-two performances.
Unsupervised Identification of Disease Marker Candidates in Retinal OCT Imaging Data
Oct 31, 2018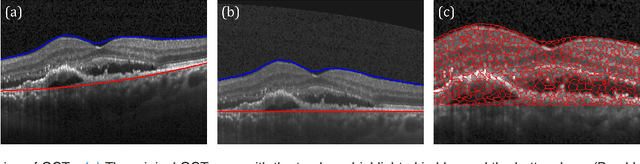
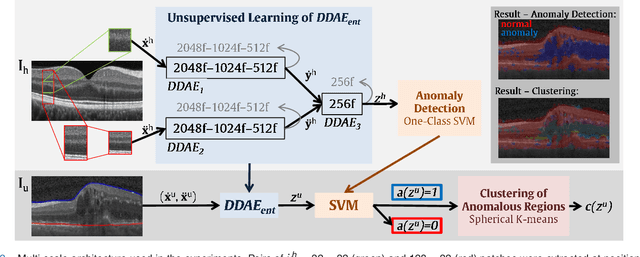
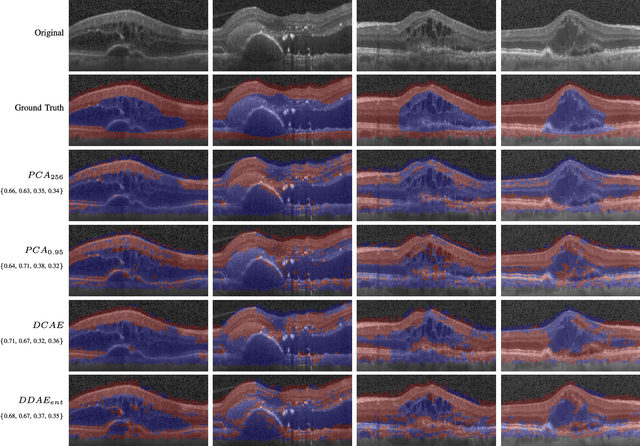
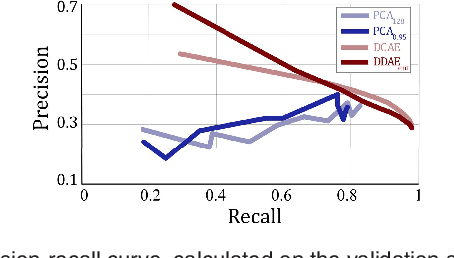
Abstract:The identification and quantification of markers in medical images is critical for diagnosis, prognosis, and disease management. Supervised machine learning enables the detection and exploitation of findings that are known a priori after annotation of training examples by experts. However, supervision does not scale well, due to the amount of necessary training examples, and the limitation of the marker vocabulary to known entities. In this proof-of-concept study, we propose unsupervised identification of anomalies as candidates for markers in retinal Optical Coherence Tomography (OCT) imaging data without a constraint to a priori definitions. We identify and categorize marker candidates occurring frequently in the data, and demonstrate that these markers show predictive value in the task of detecting disease. A careful qualitative analysis of the identified data driven markers reveals how their quantifiable occurrence aligns with our current understanding of disease course, in early- and late age-related macular degeneration (AMD) patients. A multi-scale deep denoising autoencoder is trained on healthy images, and a one-class support vector machine identifies anomalies in new data. Clustering in the anomalies identifies stable categories. Using these markers to classify healthy-, early AMD- and late AMD cases yields an accuracy of 81.40%. In a second binary classification experiment on a publicly available data set (healthy vs. intermediate AMD) the model achieves an area under the ROC curve of 0.944.
Is the winner really the best? A critical analysis of common research practice in biomedical image analysis competitions
Jun 06, 2018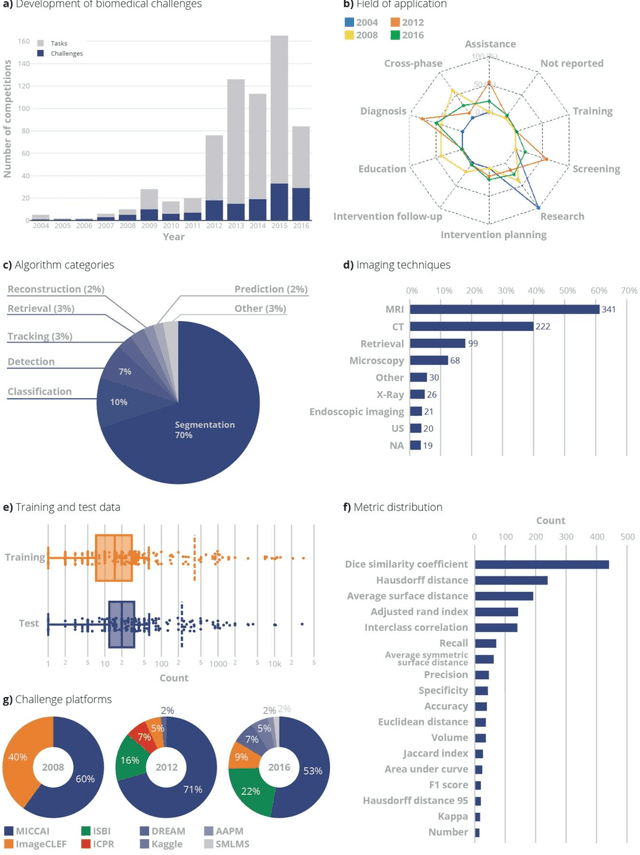
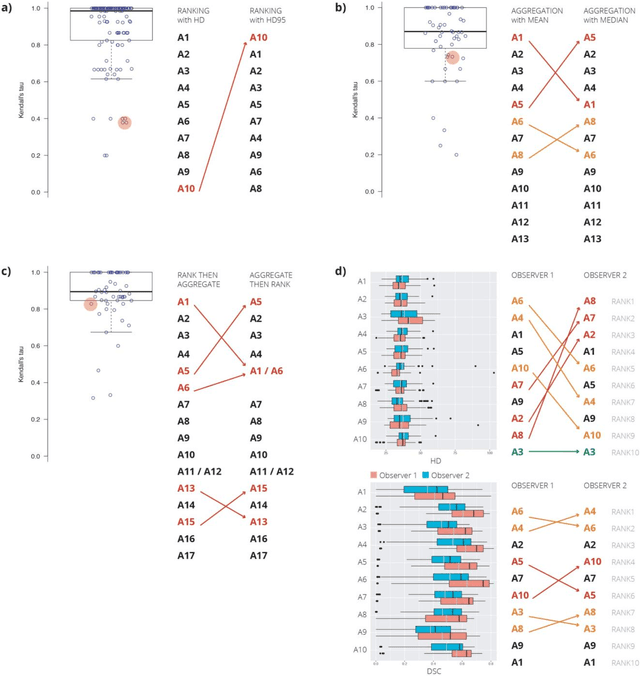
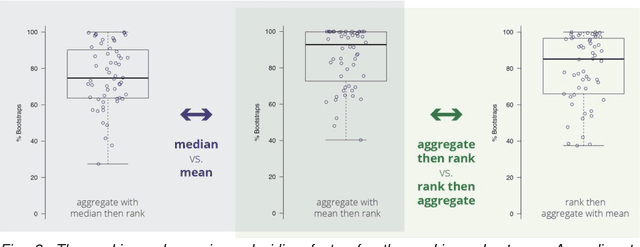
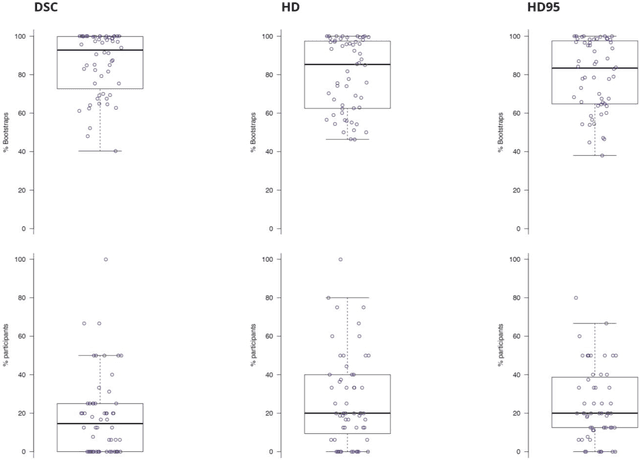
Abstract:International challenges have become the standard for validation of biomedical image analysis methods. Given their scientific impact, it is surprising that a critical analysis of common practices related to the organization of challenges has not yet been performed. In this paper, we present a comprehensive analysis of biomedical image analysis challenges conducted up to now. We demonstrate the importance of challenges and show that the lack of quality control has critical consequences. First, reproducibility and interpretation of the results is often hampered as only a fraction of relevant information is typically provided. Second, the rank of an algorithm is generally not robust to a number of variables such as the test data used for validation, the ranking scheme applied and the observers that make the reference annotations. To overcome these problems, we recommend best practice guidelines and define open research questions to be addressed in the future.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge