Taha Emre
Stochastic Siamese MAE Pretraining for Longitudinal Medical Images
Dec 29, 2025Abstract:Temporally aware image representations are crucial for capturing disease progression in 3D volumes of longitudinal medical datasets. However, recent state-of-the-art self-supervised learning approaches like Masked Autoencoding (MAE), despite their strong representation learning capabilities, lack temporal awareness. In this paper, we propose STAMP (Stochastic Temporal Autoencoder with Masked Pretraining), a Siamese MAE framework that encodes temporal information through a stochastic process by conditioning on the time difference between the 2 input volumes. Unlike deterministic Siamese approaches, which compare scans from different time points but fail to account for the inherent uncertainty in disease evolution, STAMP learns temporal dynamics stochastically by reframing the MAE reconstruction loss as a conditional variational inference objective. We evaluated STAMP on two OCT and one MRI datasets with multiple visits per patient. STAMP pretrained ViT models outperformed both existing temporal MAE methods and foundation models on different late stage Age-Related Macular Degeneration and Alzheimer's Disease progression prediction which require models to learn the underlying non-deterministic temporal dynamics of the diseases.
MIRAGE: Multimodal foundation model and benchmark for comprehensive retinal OCT image analysis
Jun 11, 2025Abstract:Artificial intelligence (AI) has become a fundamental tool for assisting clinicians in analyzing ophthalmic images, such as optical coherence tomography (OCT). However, developing AI models often requires extensive annotation, and existing models tend to underperform on independent, unseen data. Foundation models (FMs), large AI models trained on vast unlabeled datasets, have shown promise in overcoming these challenges. Nonetheless, available FMs for ophthalmology lack extensive validation, especially for segmentation tasks, and focus on a single imaging modality. In this context, we propose MIRAGE, a novel multimodal FM for the analysis of OCT and scanning laser ophthalmoscopy (SLO) images. Additionally, we propose a new evaluation benchmark with OCT/SLO classification and segmentation tasks. The comparison with general and specialized FMs and segmentation methods shows the superiority of MIRAGE in both types of tasks, highlighting its suitability as a basis for the development of robust AI systems for retinal OCT image analysis. Both MIRAGE and the evaluation benchmark are publicly available: https://github.com/j-morano/MIRAGE.
Automatic detection and prediction of nAMD activity change in retinal OCT using Siamese networks and Wasserstein Distance for ordinality
Jan 24, 2025Abstract:Neovascular age-related macular degeneration (nAMD) is a leading cause of vision loss among older adults, where disease activity detection and progression prediction are critical for nAMD management in terms of timely drug administration and improving patient outcomes. Recent advancements in deep learning offer a promising solution for predicting changes in AMD from optical coherence tomography (OCT) retinal volumes. In this work, we proposed deep learning models for the two tasks of the public MARIO Challenge at MICCAI 2024, designed to detect and forecast changes in nAMD severity with longitudinal retinal OCT. For the first task, we employ a Vision Transformer (ViT) based Siamese Network to detect changes in AMD severity by comparing scan embeddings of a patient from different time points. To train a model to forecast the change after 3 months, we exploit, for the first time, an Earth Mover (Wasserstein) Distance-based loss to harness the ordinal relation within the severity change classes. Both models ranked high on the preliminary leaderboard, demonstrating that their predictive capabilities could facilitate nAMD treatment management.
Forecasting Disease Progression with Parallel Hyperplanes in Longitudinal Retinal OCT
Sep 30, 2024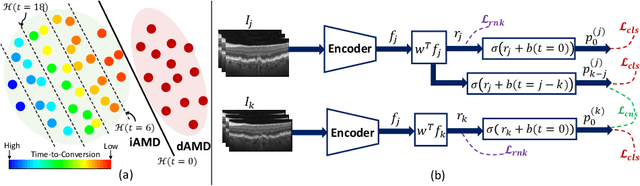
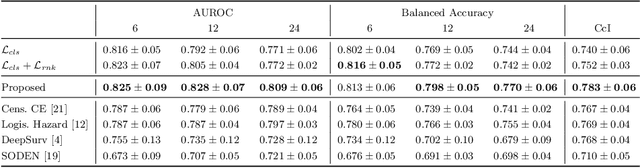
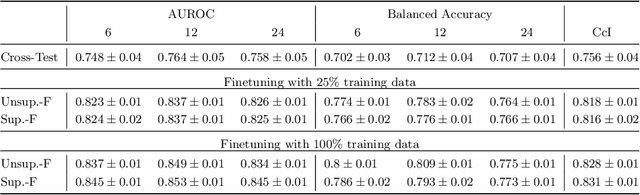
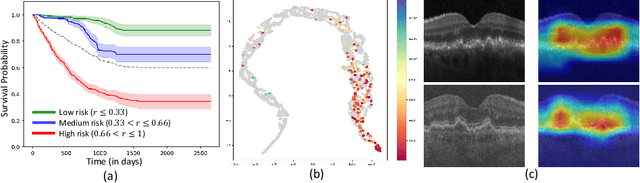
Abstract:Predicting future disease progression risk from medical images is challenging due to patient heterogeneity, and subtle or unknown imaging biomarkers. Moreover, deep learning (DL) methods for survival analysis are susceptible to image domain shifts across scanners. We tackle these issues in the task of predicting late dry Age-related Macular Degeneration (dAMD) onset from retinal OCT scans. We propose a novel DL method for survival prediction to jointly predict from the current scan a risk score, inversely related to time-to-conversion, and the probability of conversion within a time interval $t$. It uses a family of parallel hyperplanes generated by parameterizing the bias term as a function of $t$. In addition, we develop unsupervised losses based on intra-subject image pairs to ensure that risk scores increase over time and that future conversion predictions are consistent with AMD stage prediction using actual scans of future visits. Such losses enable data-efficient fine-tuning of the trained model on new unlabeled datasets acquired with a different scanner. Extensive evaluation on two large datasets acquired with different scanners resulted in a mean AUROCs of 0.82 for Dataset-1 and 0.83 for Dataset-2, across prediction intervals of 6,12 and 24 months.
Time-Equivariant Contrastive Learning for Degenerative Disease Progression in Retinal OCT
May 15, 2024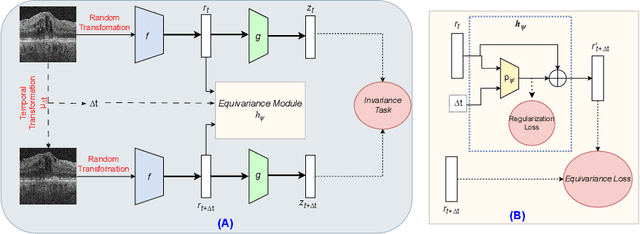
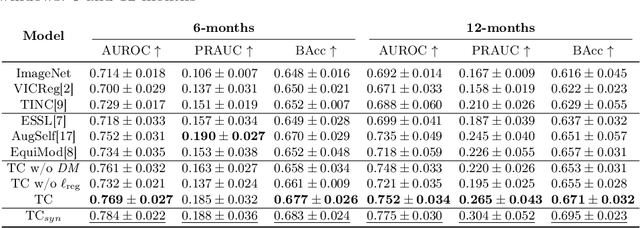
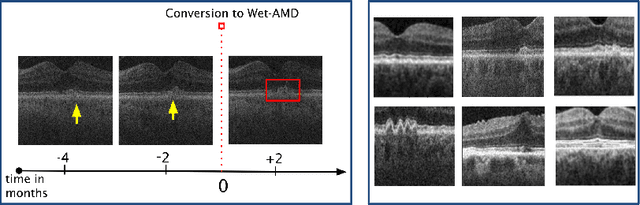
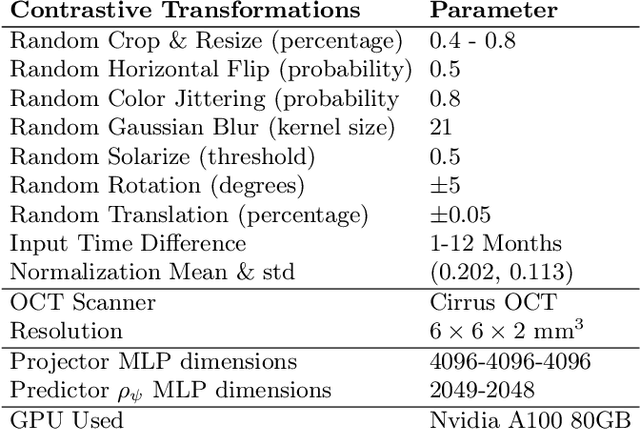
Abstract:Contrastive pretraining provides robust representations by ensuring their invariance to different image transformations while simultaneously preventing representational collapse. Equivariant contrastive learning, on the other hand, provides representations sensitive to specific image transformations while remaining invariant to others. By introducing equivariance to time-induced transformations, such as disease-related anatomical changes in longitudinal imaging, the model can effectively capture such changes in the representation space. In this work, we pro-pose a Time-equivariant Contrastive Learning (TC) method. First, an encoder embeds two unlabeled scans from different time points of the same patient into the representation space. Next, a temporal equivariance module is trained to predict the representation of a later visit based on the representation from one of the previous visits and the corresponding time interval with a novel regularization loss term while preserving the invariance property to irrelevant image transformations. On a large longitudinal dataset, our model clearly outperforms existing equivariant contrastive methods in predicting progression from intermediate age-related macular degeneration (AMD) to advanced wet-AMD within a specified time-window.
3DTINC: Time-Equivariant Non-Contrastive Learning for Predicting Disease Progression from Longitudinal OCTs
Dec 28, 2023



Abstract:Self-supervised learning (SSL) has emerged as a powerful technique for improving the efficiency and effectiveness of deep learning models. Contrastive methods are a prominent family of SSL that extract similar representations of two augmented views of an image while pushing away others in the representation space as negatives. However, the state-of-the-art contrastive methods require large batch sizes and augmentations designed for natural images that are impractical for 3D medical images. To address these limitations, we propose a new longitudinal SSL method, 3DTINC, based on non-contrastive learning. It is designed to learn perturbation-invariant features for 3D optical coherence tomography (OCT) volumes, using augmentations specifically designed for OCT. We introduce a new non-contrastive similarity loss term that learns temporal information implicitly from intra-patient scans acquired at different times. Our experiments show that this temporal information is crucial for predicting progression of retinal diseases, such as age-related macular degeneration (AMD). After pretraining with 3DTINC, we evaluated the learned representations and the prognostic models on two large-scale longitudinal datasets of retinal OCTs where we predict the conversion to wet-AMD within a six months interval. Our results demonstrate that each component of our contributions is crucial for learning meaningful representations useful in predicting disease progression from longitudinal volumetric scans.
Pretrained Deep 2.5D Models for Efficient Predictive Modeling from Retinal OCT
Jul 25, 2023Abstract:In the field of medical imaging, 3D deep learning models play a crucial role in building powerful predictive models of disease progression. However, the size of these models presents significant challenges, both in terms of computational resources and data requirements. Moreover, achieving high-quality pretraining of 3D models proves to be even more challenging. To address these issues, hybrid 2.5D approaches provide an effective solution for utilizing 3D volumetric data efficiently using 2D models. Combining 2D and 3D techniques offers a promising avenue for optimizing performance while minimizing memory requirements. In this paper, we explore 2.5D architectures based on a combination of convolutional neural networks (CNNs), long short-term memory (LSTM), and Transformers. In addition, leveraging the benefits of recent non-contrastive pretraining approaches in 2D, we enhanced the performance and data efficiency of 2.5D techniques even further. We demonstrate the effectiveness of architectures and associated pretraining on a task of predicting progression to wet age-related macular degeneration (AMD) within a six-month period on two large longitudinal OCT datasets.
Transformer-based end-to-end classification of variable-length volumetric data
Jul 21, 2023



Abstract:The automatic classification of 3D medical data is memory-intensive. Also, variations in the number of slices between samples is common. Na\"ive solutions such as subsampling can solve these problems, but at the cost of potentially eliminating relevant diagnosis information. Transformers have shown promising performance for sequential data analysis. However, their application for long sequences is data, computationally, and memory demanding. In this paper, we propose an end-to-end Transformer-based framework that allows to classify volumetric data of variable length in an efficient fashion. Particularly, by randomizing the input volume-wise resolution(#slices) during training, we enhance the capacity of the learnable positional embedding assigned to each volume slice. Consequently, the accumulated positional information in each positional embedding can be generalized to the neighbouring slices, even for high-resolution volumes at the test time. By doing so, the model will be more robust to variable volume length and amenable to different computational budgets. We evaluated the proposed approach in retinal OCT volume classification and achieved 21.96% average improvement in balanced accuracy on a 9-class diagnostic task, compared to state-of-the-art video transformers. Our findings show that varying the volume-wise resolution of the input during training results in more informative volume representation as compared to training with fixed number of slices per volume.
Morph-SSL: Self-Supervision with Longitudinal Morphing to Predict AMD Progression from OCT
Apr 17, 2023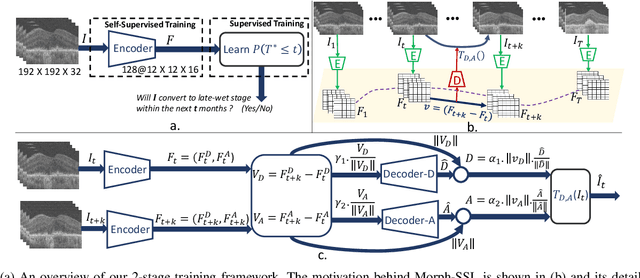
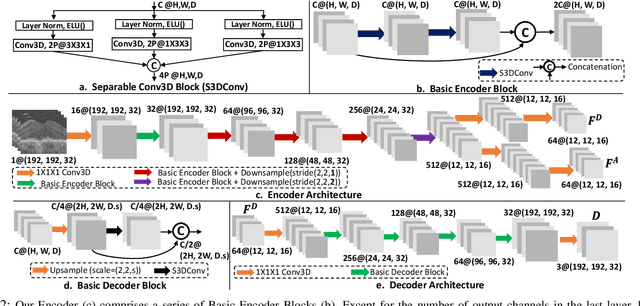

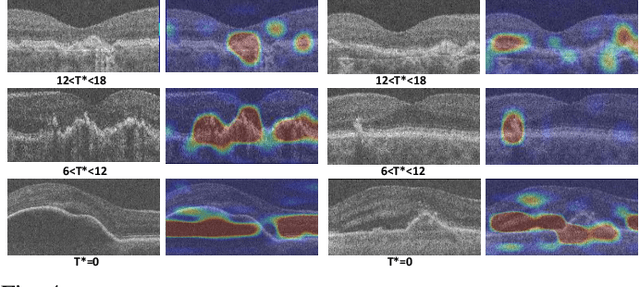
Abstract:The lack of reliable biomarkers makes predicting the conversion from intermediate to neovascular age-related macular degeneration (iAMD, nAMD) a challenging task. We develop a Deep Learning (DL) model to predict the future risk of conversion of an eye from iAMD to nAMD from its current OCT scan. Although eye clinics generate vast amounts of longitudinal OCT scans to monitor AMD progression, only a small subset can be manually labeled for supervised DL. To address this issue, we propose Morph-SSL, a novel Self-supervised Learning (SSL) method for longitudinal data. It uses pairs of unlabelled OCT scans from different visits and involves morphing the scan from the previous visit to the next. The Decoder predicts the transformation for morphing and ensures a smooth feature manifold that can generate intermediate scans between visits through linear interpolation. Next, the Morph-SSL trained features are input to a Classifier which is trained in a supervised manner to model the cumulative probability distribution of the time to conversion with a sigmoidal function. Morph-SSL was trained on unlabelled scans of 399 eyes (3570 visits). The Classifier was evaluated with a five-fold cross-validation on 2418 scans from 343 eyes with clinical labels of the conversion date. The Morph-SSL features achieved an AUC of 0.766 in predicting the conversion to nAMD within the next 6 months, outperforming the same network when trained end-to-end from scratch or pre-trained with popular SSL methods. Automated prediction of the future risk of nAMD onset can enable timely treatment and individualized AMD management.
TINC: Temporally Informed Non-Contrastive Learning for Disease Progression Modeling in Retinal OCT Volumes
Jun 30, 2022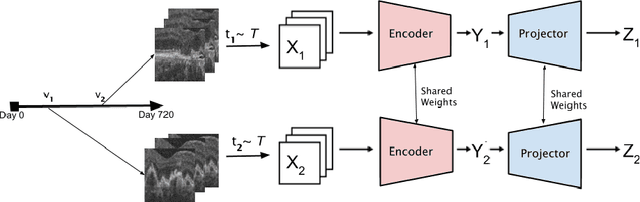
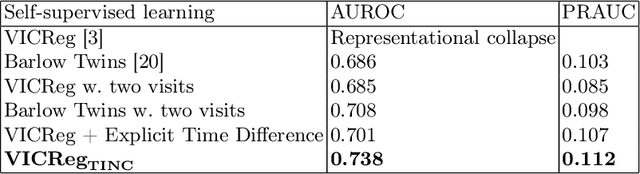

Abstract:Recent contrastive learning methods achieved state-of-the-art in low label regimes. However, the training requires large batch sizes and heavy augmentations to create multiple views of an image. With non-contrastive methods, the negatives are implicitly incorporated in the loss, allowing different images and modalities as pairs. Although the meta-information (i.e., age, sex) in medical imaging is abundant, the annotations are noisy and prone to class imbalance. In this work, we exploited already existing temporal information (different visits from a patient) in a longitudinal optical coherence tomography (OCT) dataset using temporally informed non-contrastive loss (TINC) without increasing complexity and need for negative pairs. Moreover, our novel pair-forming scheme can avoid heavy augmentations and implicitly incorporates the temporal information in the pairs. Finally, these representations learned from the pretraining are more successful in predicting disease progression where the temporal information is crucial for the downstream task. More specifically, our model outperforms existing models in predicting the risk of conversion within a time frame from intermediate age-related macular degeneration (AMD) to the late wet-AMD stage.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge