Lars Fritsche
Forecasting Disease Progression with Parallel Hyperplanes in Longitudinal Retinal OCT
Sep 30, 2024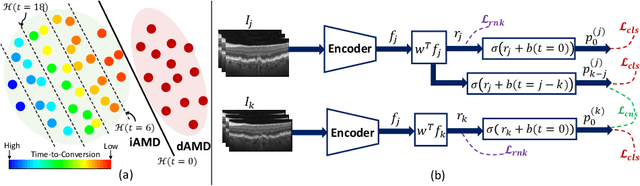
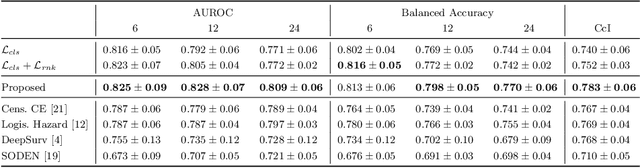
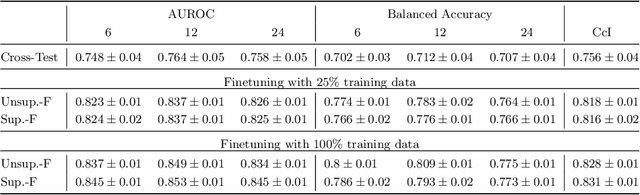
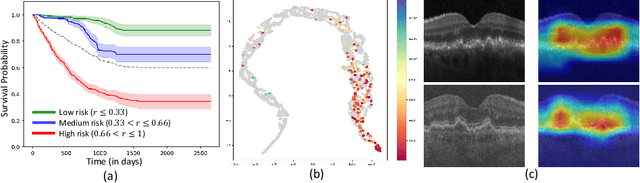
Abstract:Predicting future disease progression risk from medical images is challenging due to patient heterogeneity, and subtle or unknown imaging biomarkers. Moreover, deep learning (DL) methods for survival analysis are susceptible to image domain shifts across scanners. We tackle these issues in the task of predicting late dry Age-related Macular Degeneration (dAMD) onset from retinal OCT scans. We propose a novel DL method for survival prediction to jointly predict from the current scan a risk score, inversely related to time-to-conversion, and the probability of conversion within a time interval $t$. It uses a family of parallel hyperplanes generated by parameterizing the bias term as a function of $t$. In addition, we develop unsupervised losses based on intra-subject image pairs to ensure that risk scores increase over time and that future conversion predictions are consistent with AMD stage prediction using actual scans of future visits. Such losses enable data-efficient fine-tuning of the trained model on new unlabeled datasets acquired with a different scanner. Extensive evaluation on two large datasets acquired with different scanners resulted in a mean AUROCs of 0.82 for Dataset-1 and 0.83 for Dataset-2, across prediction intervals of 6,12 and 24 months.
Clustering disease trajectories in contrastive feature space for biomarker discovery in age-related macular degeneration
Jan 11, 2023Abstract:Age-related macular degeneration (AMD) is the leading cause of blindness in the elderly. Despite this, the exact dynamics of disease progression are poorly understood. There is a clear need for imaging biomarkers in retinal optical coherence tomography (OCT) that aid the diagnosis, prognosis and management of AMD. However, current grading systems, which coarsely group disease stage into broad categories describing early and intermediate AMD, have very limited prognostic value for the conversion to late AMD. In this paper, we are the first to analyse disease progression as clustered trajectories in a self-supervised feature space. Our method first pretrains an encoder with contrastive learning to project images from longitudinal time series to points in feature space. This enables the creation of disease trajectories, which are then denoised, partitioned and grouped into clusters. These clusters, found in two datasets containing time series of 7,912 patients imaged over eight years, were correlated with known OCT biomarkers. This reinforced efforts by four expert ophthalmologists to investigate clusters, during a clinical comparison and interpretation task, as candidates for time-dependent biomarkers that describe progression of AMD.
Metadata-enhanced contrastive learning from retinal optical coherence tomography images
Aug 04, 2022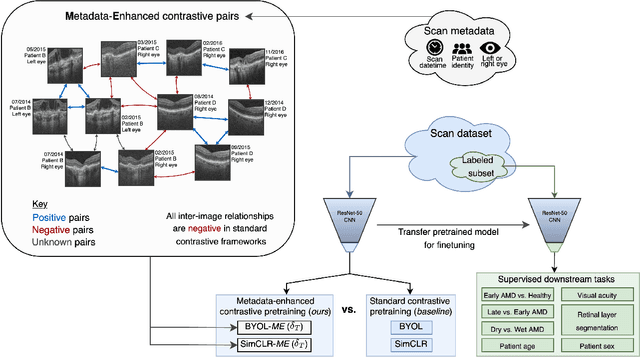
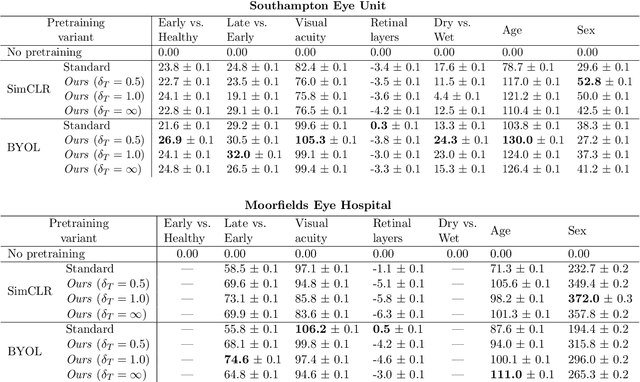
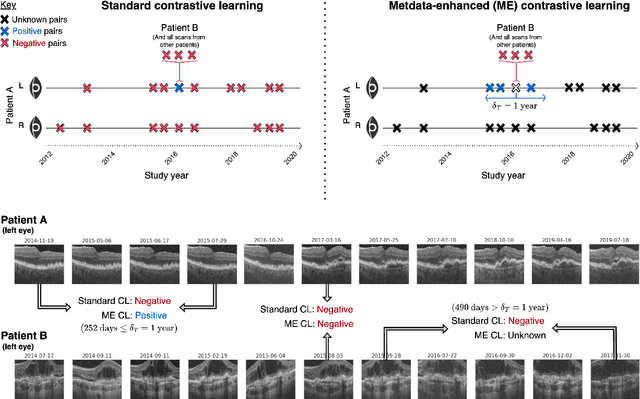
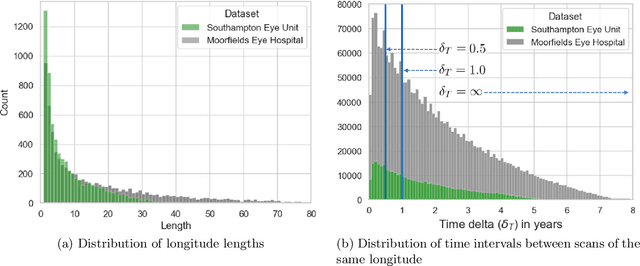
Abstract:Supervised deep learning algorithms hold great potential to automate screening, monitoring and grading of medical images. However, training performant models has typically required vast quantities of labelled data, which is scarcely available in the medical domain. Self-supervised contrastive frameworks relax this dependency by first learning from unlabelled images. In this work we show that pretraining with two contrastive methods, SimCLR and BYOL, improves the utility of deep learning with regard to the clinical assessment of age-related macular degeneration (AMD). In experiments using two large clinical datasets containing 170,427 optical coherence tomography (OCT) images of 7,912 patients, we evaluate benefits attributed to pretraining across seven downstream tasks ranging from AMD stage and type classification to prediction of functional endpoints to segmentation of retinal layers, finding performance significantly increased in six out of seven tasks with fewer labels. However, standard contrastive frameworks have two known weaknesses that are detrimental to pretraining in the medical domain. Several of the image transformations used to create positive contrastive pairs are not applicable to greyscale medical scans. Furthermore, medical images often depict the same anatomical region and disease severity, resulting in numerous misleading negative pairs. To address these issues we develop a novel metadata-enhanced approach that exploits the rich set of inherently available patient information. To this end we employ records for patient identity, eye position (i.e. left or right) and time series data to indicate the typically unknowable set of inter-image contrastive relationships. By leveraging this often neglected information our metadata-enhanced contrastive pretraining leads to further benefits and outperforms conventional contrastive methods in five out of seven downstream tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge