Hendrik Scholl
Stochastic Siamese MAE Pretraining for Longitudinal Medical Images
Dec 29, 2025Abstract:Temporally aware image representations are crucial for capturing disease progression in 3D volumes of longitudinal medical datasets. However, recent state-of-the-art self-supervised learning approaches like Masked Autoencoding (MAE), despite their strong representation learning capabilities, lack temporal awareness. In this paper, we propose STAMP (Stochastic Temporal Autoencoder with Masked Pretraining), a Siamese MAE framework that encodes temporal information through a stochastic process by conditioning on the time difference between the 2 input volumes. Unlike deterministic Siamese approaches, which compare scans from different time points but fail to account for the inherent uncertainty in disease evolution, STAMP learns temporal dynamics stochastically by reframing the MAE reconstruction loss as a conditional variational inference objective. We evaluated STAMP on two OCT and one MRI datasets with multiple visits per patient. STAMP pretrained ViT models outperformed both existing temporal MAE methods and foundation models on different late stage Age-Related Macular Degeneration and Alzheimer's Disease progression prediction which require models to learn the underlying non-deterministic temporal dynamics of the diseases.
Forecasting Disease Progression with Parallel Hyperplanes in Longitudinal Retinal OCT
Sep 30, 2024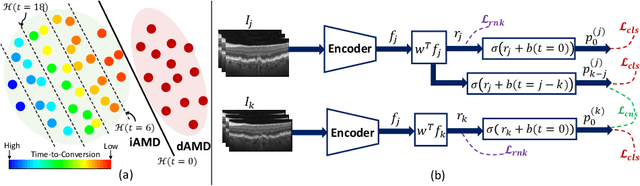
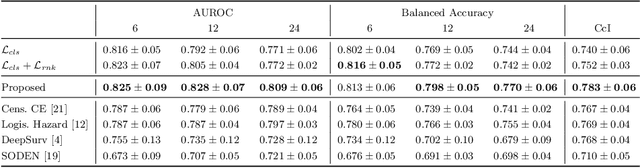
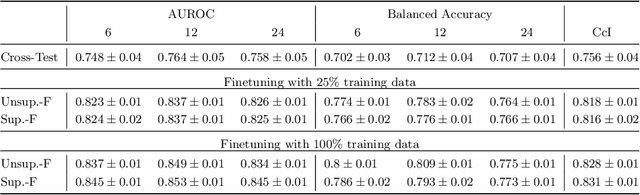
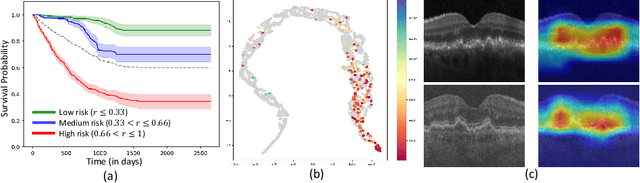
Abstract:Predicting future disease progression risk from medical images is challenging due to patient heterogeneity, and subtle or unknown imaging biomarkers. Moreover, deep learning (DL) methods for survival analysis are susceptible to image domain shifts across scanners. We tackle these issues in the task of predicting late dry Age-related Macular Degeneration (dAMD) onset from retinal OCT scans. We propose a novel DL method for survival prediction to jointly predict from the current scan a risk score, inversely related to time-to-conversion, and the probability of conversion within a time interval $t$. It uses a family of parallel hyperplanes generated by parameterizing the bias term as a function of $t$. In addition, we develop unsupervised losses based on intra-subject image pairs to ensure that risk scores increase over time and that future conversion predictions are consistent with AMD stage prediction using actual scans of future visits. Such losses enable data-efficient fine-tuning of the trained model on new unlabeled datasets acquired with a different scanner. Extensive evaluation on two large datasets acquired with different scanners resulted in a mean AUROCs of 0.82 for Dataset-1 and 0.83 for Dataset-2, across prediction intervals of 6,12 and 24 months.
Spatiotemporal Representation Learning for Short and Long Medical Image Time Series
Mar 12, 2024Abstract:Analyzing temporal developments is crucial for the accurate prognosis of many medical conditions. Temporal changes that occur over short time scales are key to assessing the health of physiological functions, such as the cardiac cycle. Moreover, tracking longer term developments that occur over months or years in evolving processes, such as age-related macular degeneration (AMD), is essential for accurate prognosis. Despite the importance of both short and long term analysis to clinical decision making, they remain understudied in medical deep learning. State of the art methods for spatiotemporal representation learning, developed for short natural videos, prioritize the detection of temporal constants rather than temporal developments. Moreover, they do not account for varying time intervals between acquisitions, which are essential for contextualizing observed changes. To address these issues, we propose two approaches. First, we combine clip-level contrastive learning with a novel temporal embedding to adapt to irregular time series. Second, we propose masking and predicting latent frame representations of the temporal sequence. Our two approaches outperform all prior methods on temporally-dependent tasks including cardiac output estimation and three prognostic AMD tasks. Overall, this enables the automated analysis of temporal patterns which are typically overlooked in applications of deep learning to medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge