Martin J. Menten
on behalf of the PINNACLE consortium
Stochastic Siamese MAE Pretraining for Longitudinal Medical Images
Dec 29, 2025Abstract:Temporally aware image representations are crucial for capturing disease progression in 3D volumes of longitudinal medical datasets. However, recent state-of-the-art self-supervised learning approaches like Masked Autoencoding (MAE), despite their strong representation learning capabilities, lack temporal awareness. In this paper, we propose STAMP (Stochastic Temporal Autoencoder with Masked Pretraining), a Siamese MAE framework that encodes temporal information through a stochastic process by conditioning on the time difference between the 2 input volumes. Unlike deterministic Siamese approaches, which compare scans from different time points but fail to account for the inherent uncertainty in disease evolution, STAMP learns temporal dynamics stochastically by reframing the MAE reconstruction loss as a conditional variational inference objective. We evaluated STAMP on two OCT and one MRI datasets with multiple visits per patient. STAMP pretrained ViT models outperformed both existing temporal MAE methods and foundation models on different late stage Age-Related Macular Degeneration and Alzheimer's Disease progression prediction which require models to learn the underlying non-deterministic temporal dynamics of the diseases.
Synthetic Vasculature and Pathology Enhance Vision-Language Model Reasoning
Dec 11, 2025Abstract:Vision-Language Models (VLMs) offer a promising path toward interpretable medical diagnosis by allowing users to ask about clinical explanations alongside predictions and across different modalities. However, training VLMs for detailed reasoning requires large-scale image-text datasets. In many specialized domains, for example in reading Optical Coherence Tomography Angiography (OCTA) images, such precise text with grounded description of pathologies is scarce or even non-existent. To overcome this bottleneck, we introduce Synthetic Vasculature Reasoning (SVR), a framework that controllably synthesizes images and corresponding text, specifically: realistic retinal vasculature with Diabetic Retinopathy (DR) features: capillary dropout, microaneurysms, neovascularization, and tortuosity, while automatically generating granular reasoning texts. Based on this we curate OCTA-100K-SVR, an OCTA image-reasoning dataset with 100,000 pairs. Our experiments show that a general-purpose VLM (Qwen3-VL-8b) trained on the dataset achieves a zero-shot balanced classification accuracy of 89.67% on real OCTA images, outperforming supervised baselines. Through human expert evaluation we also demonstrate that it significantly enhances explanation quality and pathology localization on clinical data.
Fine-tuning Vision Language Models with Graph-based Knowledge for Explainable Medical Image Analysis
Mar 12, 2025



Abstract:Accurate staging of Diabetic Retinopathy (DR) is essential for guiding timely interventions and preventing vision loss. However, current staging models are hardly interpretable, and most public datasets contain no clinical reasoning or interpretation beyond image-level labels. In this paper, we present a novel method that integrates graph representation learning with vision-language models (VLMs) to deliver explainable DR diagnosis. Our approach leverages optical coherence tomography angiography (OCTA) images by constructing biologically informed graphs that encode key retinal vascular features such as vessel morphology and spatial connectivity. A graph neural network (GNN) then performs DR staging while integrated gradients highlight critical nodes and edges and their individual features that drive the classification decisions. We collect this graph-based knowledge which attributes the model's prediction to physiological structures and their characteristics. We then transform it into textual descriptions for VLMs. We perform instruction-tuning with these textual descriptions and the corresponding image to train a student VLM. This final agent can classify the disease and explain its decision in a human interpretable way solely based on a single image input. Experimental evaluations on both proprietary and public datasets demonstrate that our method not only improves classification accuracy but also offers more clinically interpretable results. An expert study further demonstrates that our method provides more accurate diagnostic explanations and paves the way for precise localization of pathologies in OCTA images.
Skelite: Compact Neural Networks for Efficient Iterative Skeletonization
Mar 10, 2025



Abstract:Skeletonization extracts thin representations from images that compactly encode their geometry and topology. These representations have become an important topological prior for preserving connectivity in curvilinear structures, aiding medical tasks like vessel segmentation. Existing compatible skeletonization algorithms face significant trade-offs: morphology-based approaches are computationally efficient but prone to frequent breakages, while topology-preserving methods require substantial computational resources. We propose a novel framework for training iterative skeletonization algorithms with a learnable component. The framework leverages synthetic data, task-specific augmentation, and a model distillation strategy to learn compact neural networks that produce thin, connected skeletons with a fully differentiable iterative algorithm. Our method demonstrates a 100 times speedup over topology-constrained algorithms while maintaining high accuracy and generalizing effectively to new domains without fine-tuning. Benchmarking and downstream validation in 2D and 3D tasks demonstrate its computational efficiency and real-world applicability
Interpretable Retinal Disease Prediction Using Biology-Informed Heterogeneous Graph Representations
Feb 23, 2025



Abstract:Interpretability is crucial to enhance trust in machine learning models for medical diagnostics. However, most state-of-the-art image classifiers based on neural networks are not interpretable. As a result, clinicians often resort to known biomarkers for diagnosis, although biomarker-based classification typically performs worse than large neural networks. This work proposes a method that surpasses the performance of established machine learning models while simultaneously improving prediction interpretability for diabetic retinopathy staging from optical coherence tomography angiography (OCTA) images. Our method is based on a novel biology-informed heterogeneous graph representation that models retinal vessel segments, intercapillary areas, and the foveal avascular zone (FAZ) in a human-interpretable way. This graph representation allows us to frame diabetic retinopathy staging as a graph-level classification task, which we solve using an efficient graph neural network. We benchmark our method against well-established baselines, including classical biomarker-based classifiers, convolutional neural networks (CNNs), and vision transformers. Our model outperforms all baselines on two datasets. Crucially, we use our biology-informed graph to provide explanations of unprecedented detail. Our approach surpasses existing methods in precisely localizing and identifying critical vessels or intercapillary areas. In addition, we give informative and human-interpretable attributions to critical characteristics. Our work contributes to the development of clinical decision-support tools in ophthalmology.
Towards Generalisable Time Series Understanding Across Domains
Oct 09, 2024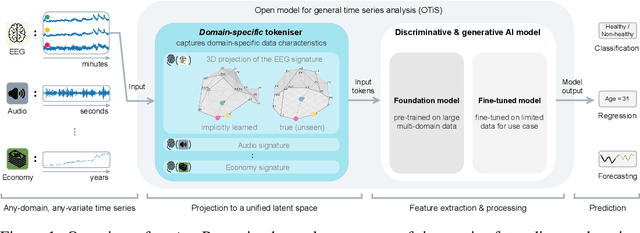
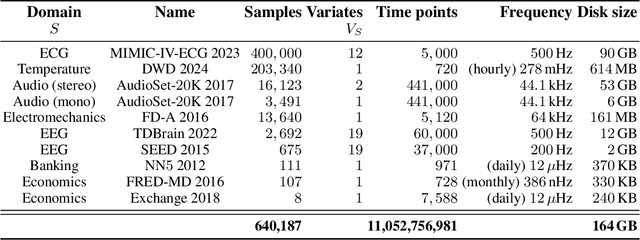
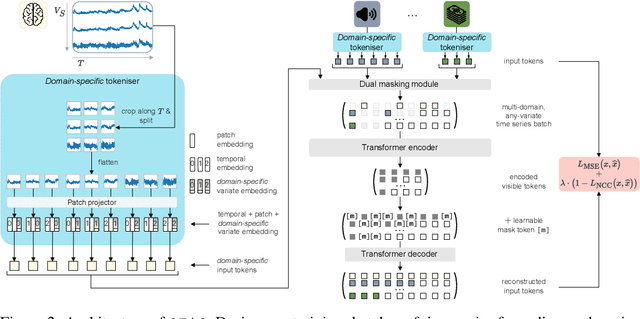
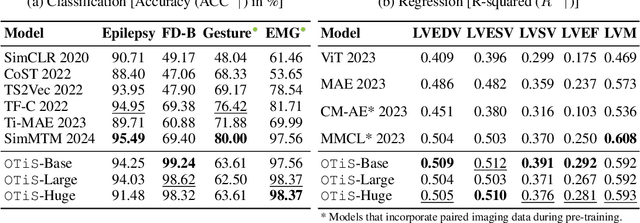
Abstract:In natural language processing and computer vision, self-supervised pre-training on large datasets unlocks foundational model capabilities across domains and tasks. However, this potential has not yet been realised in time series analysis, where existing methods disregard the heterogeneous nature of time series characteristics. Time series are prevalent in many domains, including medicine, engineering, natural sciences, and finance, but their characteristics vary significantly in terms of variate count, inter-variate relationships, temporal dynamics, and sampling frequency. This inherent heterogeneity across domains prevents effective pre-training on large time series corpora. To address this issue, we introduce OTiS, an open model for general time series analysis, that has been specifically designed to handle multi-domain heterogeneity. We propose a novel pre-training paradigm including a tokeniser with learnable domain-specific signatures, a dual masking strategy to capture temporal causality, and a normalised cross-correlation loss to model long-range dependencies. Our model is pre-trained on a large corpus of 640,187 samples and 11 billion time points spanning 8 distinct domains, enabling it to analyse time series from any (unseen) domain. In comprehensive experiments across 15 diverse applications - including classification, regression, and forecasting - OTiS showcases its ability to accurately capture domain-specific data characteristics and demonstrates its competitiveness against state-of-the-art baselines. Our code and pre-trained weights are publicly available at https://github.com/oetu/otis.
Specialist vision-language models for clinical ophthalmology
Jul 11, 2024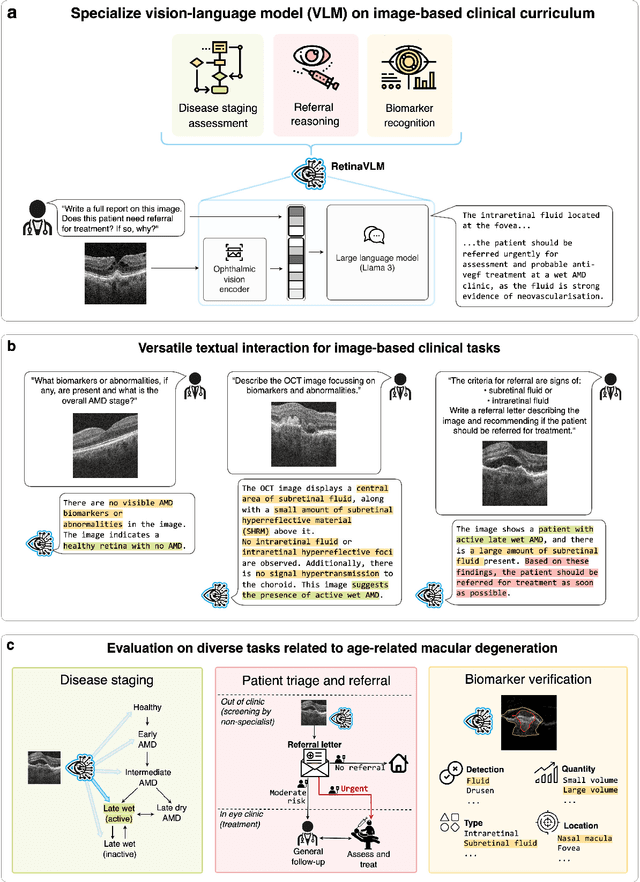

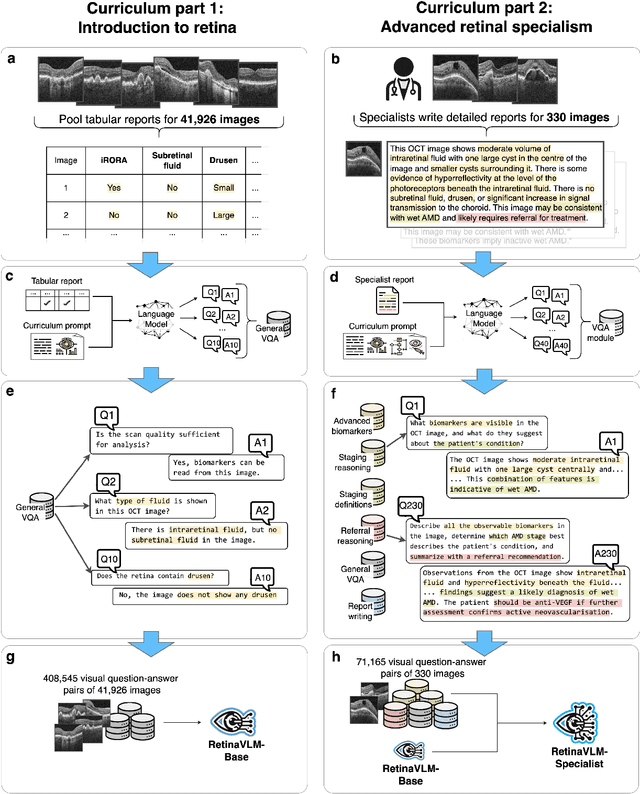

Abstract:Clinicians spend a significant amount of time reviewing medical images and transcribing their findings regarding patient diagnosis, referral and treatment in text form. Vision-language models (VLMs), which automatically interpret images and summarize their findings as text, have enormous potential to alleviate clinical workloads and increase patient access to high-quality medical care. While foundational models have stirred considerable interest in the medical community, it is unclear whether their general capabilities translate to real-world clinical utility. In this work, we show that foundation VLMs markedly underperform compared to practicing ophthalmologists on specialist tasks crucial to the care of patients with age-related macular degeneration (AMD). To address this, we initially identified the essential capabilities required for image-based clinical decision-making, and then developed a curriculum to selectively train VLMs in these skills. The resulting model, RetinaVLM, can be instructed to write reports that significantly outperform those written by leading foundation medical VLMs in disease staging (F1 score of 0.63 vs. 0.11) and patient referral (0.67 vs. 0.39), and approaches the diagnostic performance of junior ophthalmologists (who achieve 0.77 and 0.78 on the respective tasks). Furthermore, in a reader study involving two senior ophthalmologists with up to 32 years of experience, RetinaVLM's reports were found to be similarly correct (78.6% vs. 82.1%) and complete (both 78.6%) as reports written by junior ophthalmologists with up to 10 years of experience. These results demonstrate that our curriculum-based approach provides a blueprint for specializing generalist foundation medical VLMs to handle real-world clinical tasks.
Spatiotemporal Representation Learning for Short and Long Medical Image Time Series
Mar 12, 2024Abstract:Analyzing temporal developments is crucial for the accurate prognosis of many medical conditions. Temporal changes that occur over short time scales are key to assessing the health of physiological functions, such as the cardiac cycle. Moreover, tracking longer term developments that occur over months or years in evolving processes, such as age-related macular degeneration (AMD), is essential for accurate prognosis. Despite the importance of both short and long term analysis to clinical decision making, they remain understudied in medical deep learning. State of the art methods for spatiotemporal representation learning, developed for short natural videos, prioritize the detection of temporal constants rather than temporal developments. Moreover, they do not account for varying time intervals between acquisitions, which are essential for contextualizing observed changes. To address these issues, we propose two approaches. First, we combine clip-level contrastive learning with a novel temporal embedding to adapt to irregular time series. Second, we propose masking and predicting latent frame representations of the temporal sequence. Our two approaches outperform all prior methods on temporally-dependent tasks including cardiac output estimation and three prognostic AMD tasks. Overall, this enables the automated analysis of temporal patterns which are typically overlooked in applications of deep learning to medicine.
Cross-domain and Cross-dimension Learning for Image-to-Graph Transformers
Mar 11, 2024



Abstract:Direct image-to-graph transformation is a challenging task that solves object detection and relationship prediction in a single model. Due to the complexity of this task, large training datasets are rare in many domains, which makes the training of large networks challenging. This data sparsity necessitates the establishment of pre-training strategies akin to the state-of-the-art in computer vision. In this work, we introduce a set of methods enabling cross-domain and cross-dimension transfer learning for image-to-graph transformers. We propose (1) a regularized edge sampling loss for sampling the optimal number of object relationships (edges) across domains, (2) a domain adaptation framework for image-to-graph transformers that aligns features from different domains, and (3) a simple projection function that allows us to pretrain 3D transformers on 2D input data. We demonstrate our method's utility in cross-domain and cross-dimension experiments, where we pretrain our models on 2D satellite images before applying them to vastly different target domains in 2D and 3D. Our method consistently outperforms a series of baselines on challenging benchmarks, such as retinal or whole-brain vessel graph extraction.
Propagation and Attribution of Uncertainty in Medical Imaging Pipelines
Sep 28, 2023Abstract:Uncertainty estimation, which provides a means of building explainable neural networks for medical imaging applications, have mostly been studied for single deep learning models that focus on a specific task. In this paper, we propose a method to propagate uncertainty through cascades of deep learning models in medical imaging pipelines. This allows us to aggregate the uncertainty in later stages of the pipeline and to obtain a joint uncertainty measure for the predictions of later models. Additionally, we can separately report contributions of the aleatoric, data-based, uncertainty of every component in the pipeline. We demonstrate the utility of our method on a realistic imaging pipeline that reconstructs undersampled brain and knee magnetic resonance (MR) images and subsequently predicts quantitative information from the images, such as the brain volume, or knee side or patient's sex. We quantitatively show that the propagated uncertainty is correlated with input uncertainty and compare the proportions of contributions of pipeline stages to the joint uncertainty measure.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge