Markus Graf
Evaluation of GPT-4 for chest X-ray impression generation: A reader study on performance and perception
Nov 12, 2023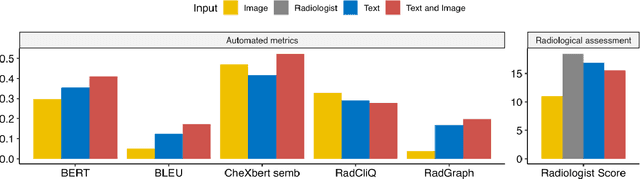
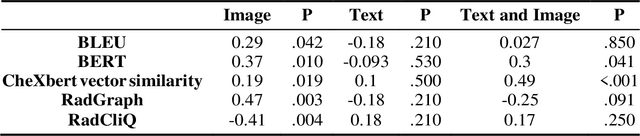
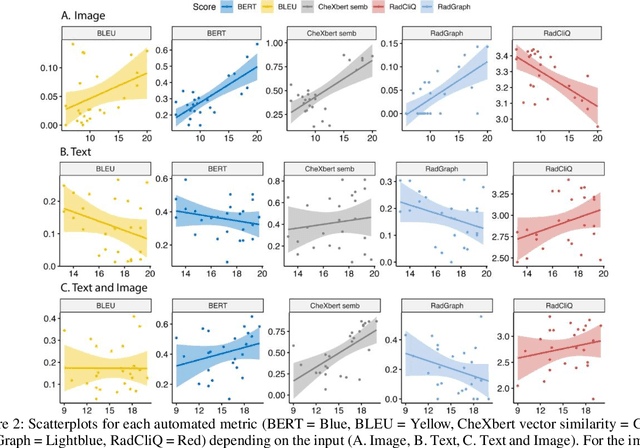
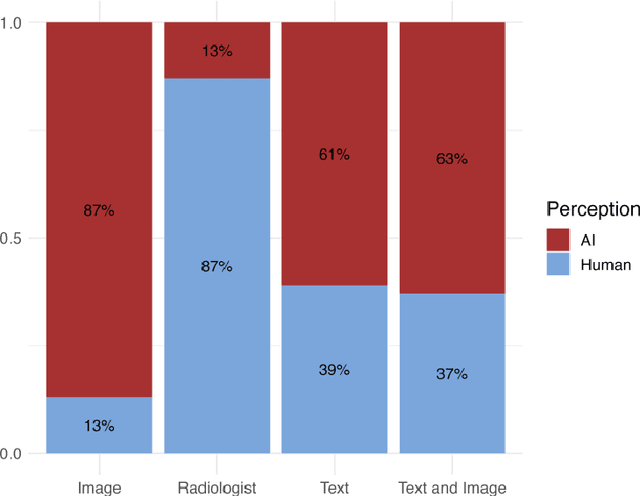
Abstract:The remarkable generative capabilities of multimodal foundation models are currently being explored for a variety of applications. Generating radiological impressions is a challenging task that could significantly reduce the workload of radiologists. In our study we explored and analyzed the generative abilities of GPT-4 for Chest X-ray impression generation. To generate and evaluate impressions of chest X-rays based on different input modalities (image, text, text and image), a blinded radiological report was written for 25-cases of the publicly available NIH-dataset. GPT-4 was given image, finding section or both sequentially to generate an input dependent impression. In a blind randomized reading, 4-radiologists rated the impressions and were asked to classify the impression origin (Human, AI), providing justification for their decision. Lastly text model evaluation metrics and their correlation with the radiological score (summation of the 4 dimensions) was assessed. According to the radiological score, the human-written impression was rated highest, although not significantly different to text-based impressions. The automated evaluation metrics showed moderate to substantial correlations to the radiological score for the image impressions, however individual scores were highly divergent among inputs, indicating insufficient representation of radiological quality. Detection of AI-generated impressions varied by input and was 61% for text-based impressions. Impressions classified as AI-generated had significantly worse radiological scores even when written by a radiologist, indicating potential bias. Our study revealed significant discrepancies between a radiological assessment and common automatic evaluation metrics depending on the model input. The detection of AI-generated findings is subject to bias that highly rated impressions are perceived as human-written.
3D Arterial Segmentation via Single 2D Projections and Depth Supervision in Contrast-Enhanced CT Images
Sep 15, 2023Abstract:Automated segmentation of the blood vessels in 3D volumes is an essential step for the quantitative diagnosis and treatment of many vascular diseases. 3D vessel segmentation is being actively investigated in existing works, mostly in deep learning approaches. However, training 3D deep networks requires large amounts of manual 3D annotations from experts, which are laborious to obtain. This is especially the case for 3D vessel segmentation, as vessels are sparse yet spread out over many slices and disconnected when visualized in 2D slices. In this work, we propose a novel method to segment the 3D peripancreatic arteries solely from one annotated 2D projection per training image with depth supervision. We perform extensive experiments on the segmentation of peripancreatic arteries on 3D contrast-enhanced CT images and demonstrate how well we capture the rich depth information from 2D projections. We demonstrate that by annotating a single, randomly chosen projection for each training sample, we obtain comparable performance to annotating multiple 2D projections, thereby reducing the annotation effort. Furthermore, by mapping the 2D labels to the 3D space using depth information and incorporating this into training, we almost close the performance gap between 3D supervision and 2D supervision. Our code is available at: https://github.com/alinafdima/3Dseg-mip-depth.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge