Mert R. Sabuncu
AI-Based Detection of In-Treatment Changes from Prostate MR-Linac Images
Feb 04, 2026Abstract:Purpose: To investigate whether routinely acquired longitudinal MR-Linac images can be leveraged to characterize treatment-induced changes during radiotherapy, particularly subtle inter-fraction changes over short intervals (average of 2 days). Materials and Methods: This retrospective study included a series of 0.35T MR-Linac images from 761 patients. An artificial intelligence (deep learning) model was used to characterize treatment-induced changes by predicting the temporal order of paired images. The model was first trained with the images from the first and the last fractions (F1-FL), then with all pairs (All-pairs). Model performance was assessed using quantitative metrics (accuracy and AUC), compared to a radiologist's performance, and qualitative analyses - the saliency map evaluation to investigate affected anatomical regions. Input ablation experiments were performed to identify the anatomical regions altered by radiotherapy. The radiologist conducted an additional task on partial images reconstructed by saliency map regions, reporting observations as well. Quantitative image analysis was conducted to investigate the results from the model and the radiologist. Results: The F1-FL model yielded near-perfect performance (AUC of 0.99), significantly outperforming the radiologist. The All-pairs model yielded an AUC of 0.97. This performance reflects therapy-induced changes, supported by the performance correlation to fraction intervals, ablation tests and expert's interpretation. Primary regions driving the predictions were prostate, bladder, and pubic symphysis. Conclusion: The model accurately predicts temporal order of MR-Linac fractions and detects radiation-induced changes over one or a few days, including prostate and adjacent organ alterations confirmed by experts. This underscores MR-Linac's potential for advanced image analysis beyond image guidance.
AtlasMorph: Learning conditional deformable templates for brain MRI
Nov 17, 2025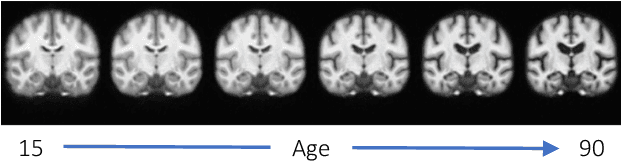
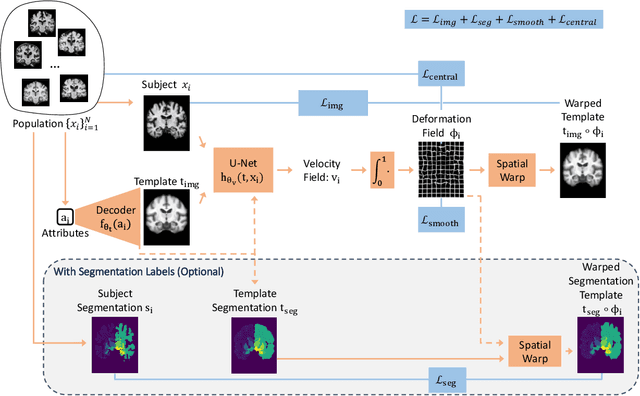
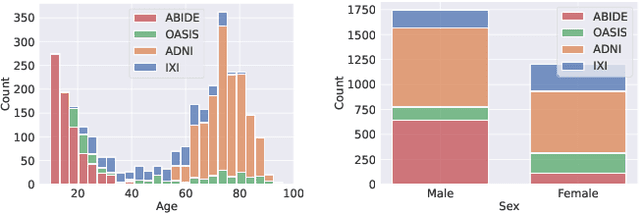
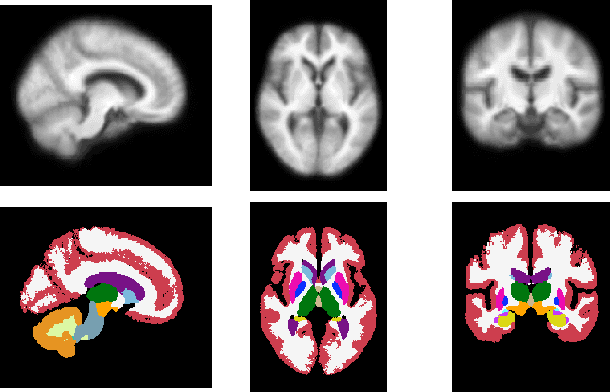
Abstract:Deformable templates, or atlases, are images that represent a prototypical anatomy for a population, and are often enhanced with probabilistic anatomical label maps. They are commonly used in medical image analysis for population studies and computational anatomy tasks such as registration and segmentation. Because developing a template is a computationally expensive process, relatively few templates are available. As a result, analysis is often conducted with sub-optimal templates that are not truly representative of the study population, especially when there are large variations within this population. We propose a machine learning framework that uses convolutional registration neural networks to efficiently learn a function that outputs templates conditioned on subject-specific attributes, such as age and sex. We also leverage segmentations, when available, to produce anatomical segmentation maps for the resulting templates. The learned network can also be used to register subject images to the templates. We demonstrate our method on a compilation of 3D brain MRI datasets, and show that it can learn high-quality templates that are representative of populations. We find that annotated conditional templates enable better registration than their unlabeled unconditional counterparts, and outperform other templates construction methods.
From Classification to Cross-Modal Understanding: Leveraging Vision-Language Models for Fine-Grained Renal Pathology
Nov 15, 2025Abstract:Fine-grained glomerular subtyping is central to kidney biopsy interpretation, but clinically valuable labels are scarce and difficult to obtain. Existing computational pathology approaches instead tend to evaluate coarse diseased classification under full supervision with image-only models, so it remains unclear how vision-language models (VLMs) should be adapted for clinically meaningful subtyping under data constraints. In this work, we model fine-grained glomerular subtyping as a clinically realistic few-shot problem and systematically evaluate both pathology-specialized and general-purpose vision-language models under this setting. We assess not only classification performance (accuracy, AUC, F1) but also the geometry of the learned representations, examining feature alignment between image and text embeddings and the separability of glomerular subtypes. By jointly analyzing shot count, model architecture and domain knowledge, and adaptation strategy, this study provides guidance for future model selection and training under real clinical data constraints. Our results indicate that pathology-specialized vision-language backbones, when paired with the vanilla fine-tuning, are the most effective starting point. Even with only 4-8 labeled examples per glomeruli subtype, these models begin to capture distinctions and show substantial gains in discrimination and calibration, though additional supervision continues to yield incremental improvements. We also find that the discrimination between positive and negative examples is as important as image-text alignment. Overall, our results show that supervision level and adaptation strategy jointly shape both diagnostic performance and multimodal structure, providing guidance for model selection, adaptation strategies, and annotation investment.
AI-assisted workflow enables rapid, high-fidelity breast cancer clinical trial eligibility prescreening
Nov 07, 2025



Abstract:Clinical trials play an important role in cancer care and research, yet participation rates remain low. We developed MSK-MATCH (Memorial Sloan Kettering Multi-Agent Trial Coordination Hub), an AI system for automated eligibility screening from clinical text. MSK-MATCH integrates a large language model with a curated oncology trial knowledge base and retrieval-augmented architecture providing explanations for all AI predictions grounded in source text. In a retrospective dataset of 88,518 clinical documents from 731 patients across six breast cancer trials, MSK-MATCH automatically resolved 61.9% of cases and triaged 38.1% for human review. This AI-assisted workflow achieved 98.6% accuracy, 98.4% sensitivity, and 98.7% specificity for patient-level eligibility classification, matching or exceeding performance of the human-only and AI-only comparisons. For the triaged cases requiring manual review, prepopulating eligibility screens with AI-generated explanations reduced screening time from 20 minutes to 43 seconds at an average cost of $0.96 per patient-trial pair.
M^3-GloDets: Multi-Region and Multi-Scale Analysis of Fine-Grained Diseased Glomerular Detection
Aug 25, 2025Abstract:Accurate detection of diseased glomeruli is fundamental to progress in renal pathology and underpins the delivery of reliable clinical diagnoses. Although recent advances in computer vision have produced increasingly sophisticated detection algorithms, the majority of research efforts have focused on normal glomeruli or instances of global sclerosis, leaving the wider spectrum of diseased glomerular subtypes comparatively understudied. This disparity is not without consequence; the nuanced and highly variable morphological characteristics that define these disease variants frequently elude even the most advanced computational models. Moreover, ongoing debate surrounds the choice of optimal imaging magnifications and region-of-view dimensions for fine-grained glomerular analysis, adding further complexity to the pursuit of accurate classification and robust segmentation. To bridge these gaps, we present M^3-GloDet, a systematic framework designed to enable thorough evaluation of detection models across a broad continuum of regions, scales, and classes. Within this framework, we evaluate both long-standing benchmark architectures and recently introduced state-of-the-art models that have achieved notable performance, using an experimental design that reflects the diversity of region-of-interest sizes and imaging resolutions encountered in routine digital renal pathology. As the results, we found that intermediate patch sizes offered the best balance between context and efficiency. Additionally, moderate magnifications enhanced generalization by reducing overfitting. Through systematic comparison of these approaches on a multi-class diseased glomerular dataset, our aim is to advance the understanding of model strengths and limitations, and to offer actionable insights for the refinement of automated detection strategies and clinical workflows in the digital pathology domain.
DyMorph-B2I: Dynamic and Morphology-Guided Binary-to-Instance Segmentation for Renal Pathology
Aug 21, 2025Abstract:Accurate morphological quantification of renal pathology functional units relies on instance-level segmentation, yet most existing datasets and automated methods provide only binary (semantic) masks, limiting the precision of downstream analyses. Although classical post-processing techniques such as watershed, morphological operations, and skeletonization, are often used to separate semantic masks into instances, their individual effectiveness is constrained by the diverse morphologies and complex connectivity found in renal tissue. In this study, we present DyMorph-B2I, a dynamic, morphology-guided binary-to-instance segmentation pipeline tailored for renal pathology. Our approach integrates watershed, skeletonization, and morphological operations within a unified framework, complemented by adaptive geometric refinement and customizable hyperparameter tuning for each class of functional unit. Through systematic parameter optimization, DyMorph-B2I robustly separates adherent and heterogeneous structures present in binary masks. Experimental results demonstrate that our method outperforms individual classical approaches and na\"ive combinations, enabling superior instance separation and facilitating more accurate morphometric analysis in renal pathology workflows. The pipeline is publicly available at: https://github.com/ddrrnn123/DyMorph-B2I.
RealKeyMorph: Keypoints in Real-world Coordinates for Resolution-agnostic Image Registration
Jun 12, 2025Abstract:Many real-world settings require registration of a pair of medical images that differ in spatial resolution, which may arise from differences in image acquisition parameters like pixel spacing, slice thickness, and field-of-view. However, all previous machine learning-based registration techniques resample images onto a fixed resolution. This is suboptimal because resampling can introduce artifacts due to interpolation. To address this, we present RealKeyMorph (RKM), a resolution-agnostic method for image registration. RKM is an extension of KeyMorph, a registration framework which works by training a network to learn corresponding keypoints for a given pair of images, after which a closed-form keypoint matching step is used to derive the transformation that aligns them. To avoid resampling and enable operating on the raw data, RKM outputs keypoints in real-world coordinates of the scanner. To do this, we leverage the affine matrix produced by the scanner (e.g., MRI machine) that encodes the mapping from voxel coordinates to real world coordinates. By transforming keypoints into real-world space and integrating this into the training process, RKM effectively enables the extracted keypoints to be resolution-agnostic. In our experiments, we demonstrate the advantages of RKM on the registration task for orthogonal 2D stacks of abdominal MRIs, as well as 3D volumes with varying resolutions in brain datasets.
IRS: Incremental Relationship-guided Segmentation for Digital Pathology
May 28, 2025Abstract:Continual learning is rapidly emerging as a key focus in computer vision, aiming to develop AI systems capable of continuous improvement, thereby enhancing their value and practicality in diverse real-world applications. In healthcare, continual learning holds great promise for continuously acquired digital pathology data, which is collected in hospitals on a daily basis. However, panoramic segmentation on digital whole slide images (WSIs) presents significant challenges, as it is often infeasible to obtain comprehensive annotations for all potential objects, spanning from coarse structures (e.g., regions and unit objects) to fine structures (e.g., cells). This results in temporally and partially annotated data, posing a major challenge in developing a holistic segmentation framework. Moreover, an ideal segmentation model should incorporate new phenotypes, unseen diseases, and diverse populations, making this task even more complex. In this paper, we introduce a novel and unified Incremental Relationship-guided Segmentation (IRS) learning scheme to address temporally acquired, partially annotated data while maintaining out-of-distribution (OOD) continual learning capacity in digital pathology. The key innovation of IRS lies in its ability to realize a new spatial-temporal OOD continual learning paradigm by mathematically modeling anatomical relationships between existing and newly introduced classes through a simple incremental universal proposition matrix. Experimental results demonstrate that the IRS method effectively handles the multi-scale nature of pathological segmentation, enabling precise kidney segmentation across various structures (regions, units, and cells) as well as OOD disease lesions at multiple magnifications. This capability significantly enhances domain generalization, making IRS a robust approach for real-world digital pathology applications.
Fine-tuning Vision Language Models with Graph-based Knowledge for Explainable Medical Image Analysis
Mar 12, 2025



Abstract:Accurate staging of Diabetic Retinopathy (DR) is essential for guiding timely interventions and preventing vision loss. However, current staging models are hardly interpretable, and most public datasets contain no clinical reasoning or interpretation beyond image-level labels. In this paper, we present a novel method that integrates graph representation learning with vision-language models (VLMs) to deliver explainable DR diagnosis. Our approach leverages optical coherence tomography angiography (OCTA) images by constructing biologically informed graphs that encode key retinal vascular features such as vessel morphology and spatial connectivity. A graph neural network (GNN) then performs DR staging while integrated gradients highlight critical nodes and edges and their individual features that drive the classification decisions. We collect this graph-based knowledge which attributes the model's prediction to physiological structures and their characteristics. We then transform it into textual descriptions for VLMs. We perform instruction-tuning with these textual descriptions and the corresponding image to train a student VLM. This final agent can classify the disease and explain its decision in a human interpretable way solely based on a single image input. Experimental evaluations on both proprietary and public datasets demonstrate that our method not only improves classification accuracy but also offers more clinically interpretable results. An expert study further demonstrates that our method provides more accurate diagnostic explanations and paves the way for precise localization of pathologies in OCTA images.
Cancer Type, Stage and Prognosis Assessment from Pathology Reports using LLMs
Mar 03, 2025Abstract:Large Language Models (LLMs) have shown significant promise across various natural language processing tasks. However, their application in the field of pathology, particularly for extracting meaningful insights from unstructured medical texts such as pathology reports, remains underexplored and not well quantified. In this project, we leverage state-of-the-art language models, including the GPT family, Mistral models, and the open-source Llama models, to evaluate their performance in comprehensively analyzing pathology reports. Specifically, we assess their performance in cancer type identification, AJCC stage determination, and prognosis assessment, encompassing both information extraction and higher-order reasoning tasks. Based on a detailed analysis of their performance metrics in a zero-shot setting, we developed two instruction-tuned models: Path-llama3.1-8B and Path-GPT-4o-mini-FT. These models demonstrated superior performance in zero-shot cancer type identification, staging, and prognosis assessment compared to the other models evaluated.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge