Rachit Saluja
From Classification to Cross-Modal Understanding: Leveraging Vision-Language Models for Fine-Grained Renal Pathology
Nov 15, 2025Abstract:Fine-grained glomerular subtyping is central to kidney biopsy interpretation, but clinically valuable labels are scarce and difficult to obtain. Existing computational pathology approaches instead tend to evaluate coarse diseased classification under full supervision with image-only models, so it remains unclear how vision-language models (VLMs) should be adapted for clinically meaningful subtyping under data constraints. In this work, we model fine-grained glomerular subtyping as a clinically realistic few-shot problem and systematically evaluate both pathology-specialized and general-purpose vision-language models under this setting. We assess not only classification performance (accuracy, AUC, F1) but also the geometry of the learned representations, examining feature alignment between image and text embeddings and the separability of glomerular subtypes. By jointly analyzing shot count, model architecture and domain knowledge, and adaptation strategy, this study provides guidance for future model selection and training under real clinical data constraints. Our results indicate that pathology-specialized vision-language backbones, when paired with the vanilla fine-tuning, are the most effective starting point. Even with only 4-8 labeled examples per glomeruli subtype, these models begin to capture distinctions and show substantial gains in discrimination and calibration, though additional supervision continues to yield incremental improvements. We also find that the discrimination between positive and negative examples is as important as image-text alignment. Overall, our results show that supervision level and adaptation strategy jointly shape both diagnostic performance and multimodal structure, providing guidance for model selection, adaptation strategies, and annotation investment.
Cancer Type, Stage and Prognosis Assessment from Pathology Reports using LLMs
Mar 03, 2025Abstract:Large Language Models (LLMs) have shown significant promise across various natural language processing tasks. However, their application in the field of pathology, particularly for extracting meaningful insights from unstructured medical texts such as pathology reports, remains underexplored and not well quantified. In this project, we leverage state-of-the-art language models, including the GPT family, Mistral models, and the open-source Llama models, to evaluate their performance in comprehensively analyzing pathology reports. Specifically, we assess their performance in cancer type identification, AJCC stage determination, and prognosis assessment, encompassing both information extraction and higher-order reasoning tasks. Based on a detailed analysis of their performance metrics in a zero-shot setting, we developed two instruction-tuned models: Path-llama3.1-8B and Path-GPT-4o-mini-FT. These models demonstrated superior performance in zero-shot cancer type identification, staging, and prognosis assessment compared to the other models evaluated.
The 2024 Brain Tumor Segmentation (BraTS) Challenge: Glioma Segmentation on Post-treatment MRI
May 28, 2024
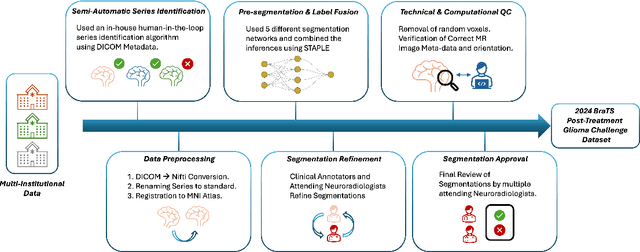
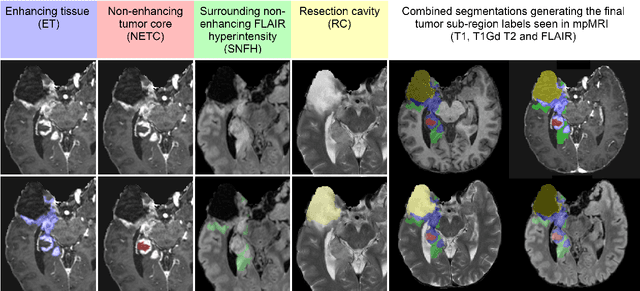
Abstract:Gliomas are the most common malignant primary brain tumors in adults and one of the deadliest types of cancer. There are many challenges in treatment and monitoring due to the genetic diversity and high intrinsic heterogeneity in appearance, shape, histology, and treatment response. Treatments include surgery, radiation, and systemic therapies, with magnetic resonance imaging (MRI) playing a key role in treatment planning and post-treatment longitudinal assessment. The 2024 Brain Tumor Segmentation (BraTS) challenge on post-treatment glioma MRI will provide a community standard and benchmark for state-of-the-art automated segmentation models based on the largest expert-annotated post-treatment glioma MRI dataset. Challenge competitors will develop automated segmentation models to predict four distinct tumor sub-regions consisting of enhancing tissue (ET), surrounding non-enhancing T2/fluid-attenuated inversion recovery (FLAIR) hyperintensity (SNFH), non-enhancing tumor core (NETC), and resection cavity (RC). Models will be evaluated on separate validation and test datasets using standardized performance metrics utilized across the BraTS 2024 cluster of challenges, including lesion-wise Dice Similarity Coefficient and Hausdorff Distance. Models developed during this challenge will advance the field of automated MRI segmentation and contribute to their integration into clinical practice, ultimately enhancing patient care.
Brain Tumor Segmentation (BraTS) Challenge 2024: Meningioma Radiotherapy Planning Automated Segmentation
May 28, 2024Abstract:The 2024 Brain Tumor Segmentation Meningioma Radiotherapy (BraTS-MEN-RT) challenge aims to advance automated segmentation algorithms using the largest known multi-institutional dataset of radiotherapy planning brain MRIs with expert-annotated target labels for patients with intact or post-operative meningioma that underwent either conventional external beam radiotherapy or stereotactic radiosurgery. Each case includes a defaced 3D post-contrast T1-weighted radiotherapy planning MRI in its native acquisition space, accompanied by a single-label "target volume" representing the gross tumor volume (GTV) and any at-risk post-operative site. Target volume annotations adhere to established radiotherapy planning protocols, ensuring consistency across cases and institutions. For pre-operative meningiomas, the target volume encompasses the entire GTV and associated nodular dural tail, while for post-operative cases, it includes at-risk resection cavity margins as determined by the treating institution. Case annotations were reviewed and approved by expert neuroradiologists and radiation oncologists. Participating teams will develop, containerize, and evaluate automated segmentation models using this comprehensive dataset. Model performance will be assessed using the lesion-wise Dice Similarity Coefficient and the 95% Hausdorff distance. The top-performing teams will be recognized at the Medical Image Computing and Computer Assisted Intervention Conference in October 2024. BraTS-MEN-RT is expected to significantly advance automated radiotherapy planning by enabling precise tumor segmentation and facilitating tailored treatment, ultimately improving patient outcomes.
BrainMorph: A Foundational Keypoint Model for Robust and Flexible Brain MRI Registration
May 22, 2024Abstract:We present a keypoint-based foundation model for general purpose brain MRI registration, based on the recently-proposed KeyMorph framework. Our model, called BrainMorph, serves as a tool that supports multi-modal, pairwise, and scalable groupwise registration. BrainMorph is trained on a massive dataset of over 100,000 3D volumes, skull-stripped and non-skull-stripped, from nearly 16,000 unique healthy and diseased subjects. BrainMorph is robust to large misalignments, interpretable via interrogating automatically-extracted keypoints, and enables rapid and controllable generation of many plausible transformations with different alignment types and different degrees of nonlinearity at test-time. We demonstrate the superiority of BrainMorph in solving 3D rigid, affine, and nonlinear registration on a variety of multi-modal brain MRI scans of healthy and diseased subjects, in both the pairwise and groupwise setting. In particular, we show registration accuracy and speeds that surpass current state-of-the-art methods, especially in the context of large initial misalignments and large group settings. All code and models are available at https://github.com/alanqrwang/brainmorph.
Analysis of the BraTS 2023 Intracranial Meningioma Segmentation Challenge
May 16, 2024



Abstract:We describe the design and results from the BraTS 2023 Intracranial Meningioma Segmentation Challenge. The BraTS Meningioma Challenge differed from prior BraTS Glioma challenges in that it focused on meningiomas, which are typically benign extra-axial tumors with diverse radiologic and anatomical presentation and a propensity for multiplicity. Nine participating teams each developed deep-learning automated segmentation models using image data from the largest multi-institutional systematically expert annotated multilabel multi-sequence meningioma MRI dataset to date, which included 1000 training set cases, 141 validation set cases, and 283 hidden test set cases. Each case included T2, T2/FLAIR, T1, and T1Gd brain MRI sequences with associated tumor compartment labels delineating enhancing tumor, non-enhancing tumor, and surrounding non-enhancing T2/FLAIR hyperintensity. Participant automated segmentation models were evaluated and ranked based on a scoring system evaluating lesion-wise metrics including dice similarity coefficient (DSC) and 95% Hausdorff Distance. The top ranked team had a lesion-wise median dice similarity coefficient (DSC) of 0.976, 0.976, and 0.964 for enhancing tumor, tumor core, and whole tumor, respectively and a corresponding average DSC of 0.899, 0.904, and 0.871, respectively. These results serve as state-of-the-art benchmarks for future pre-operative meningioma automated segmentation algorithms. Additionally, we found that 1286 of 1424 cases (90.3%) had at least 1 compartment voxel abutting the edge of the skull-stripped image edge, which requires further investigation into optimal pre-processing face anonymization steps.
A Framework for Interpretability in Machine Learning for Medical Imaging
Oct 02, 2023

Abstract:Interpretability for machine learning models in medical imaging (MLMI) is an important direction of research. However, there is a general sense of murkiness in what interpretability means. Why does the need for interpretability in MLMI arise? What goals does one actually seek to address when interpretability is needed? To answer these questions, we identify a need to formalize the goals and elements of interpretability in MLMI. By reasoning about real-world tasks and goals common in both medical image analysis and its intersection with machine learning, we identify four core elements of interpretability: localization, visual recognizability, physical attribution, and transparency. Overall, this paper formalizes interpretability needs in the context of medical imaging, and our applied perspective clarifies concrete MLMI-specific goals and considerations in order to guide method design and improve real-world usage. Our goal is to provide practical and didactic information for model designers and practitioners, inspire developers of models in the medical imaging field to reason more deeply about what interpretability is achieving, and suggest future directions of interpretability research.
The Brain Tumor Segmentation (BraTS-METS) Challenge 2023: Brain Metastasis Segmentation on Pre-treatment MRI
Jun 01, 2023



Abstract:Clinical monitoring of metastatic disease to the brain can be a laborious and time-consuming process, especially in cases involving multiple metastases when the assessment is performed manually. The Response Assessment in Neuro-Oncology Brain Metastases (RANO-BM) guideline, which utilizes the unidimensional longest diameter, is commonly used in clinical and research settings to evaluate response to therapy in patients with brain metastases. However, accurate volumetric assessment of the lesion and surrounding peri-lesional edema holds significant importance in clinical decision-making and can greatly enhance outcome prediction. The unique challenge in performing segmentations of brain metastases lies in their common occurrence as small lesions. Detection and segmentation of lesions that are smaller than 10 mm in size has not demonstrated high accuracy in prior publications. The brain metastases challenge sets itself apart from previously conducted MICCAI challenges on glioma segmentation due to the significant variability in lesion size. Unlike gliomas, which tend to be larger on presentation scans, brain metastases exhibit a wide range of sizes and tend to include small lesions. We hope that the BraTS-METS dataset and challenge will advance the field of automated brain metastasis detection and segmentation.
The University of California San Francisco, Brain Metastases Stereotactic Radiosurgery (UCSF-BMSR) MRI Dataset
Apr 19, 2023Abstract:The University of California San Francisco Brain Metastases Stereotactic Radiosurgery (UCSF-BMSR) dataset is a public, clinical, multimodal brain MRI dataset consisting of 560 brain MRIs from 412 patients with expert annotations of 5136 brain metastases. Data consists of registered and skull stripped T1 post-contrast, T1 pre-contrast, FLAIR and subtraction (T1 pre-contrast - T1 post-contrast) images and voxelwise segmentations of enhancing brain metastases in NifTI format. The dataset also includes patient demographics, surgical status and primary cancer types. The UCSF-BSMR has been made publicly available in the hopes that researchers will use these data to push the boundaries of AI applications for brain metastases.
Movement science needs different pose tracking algorithms
Jul 24, 2019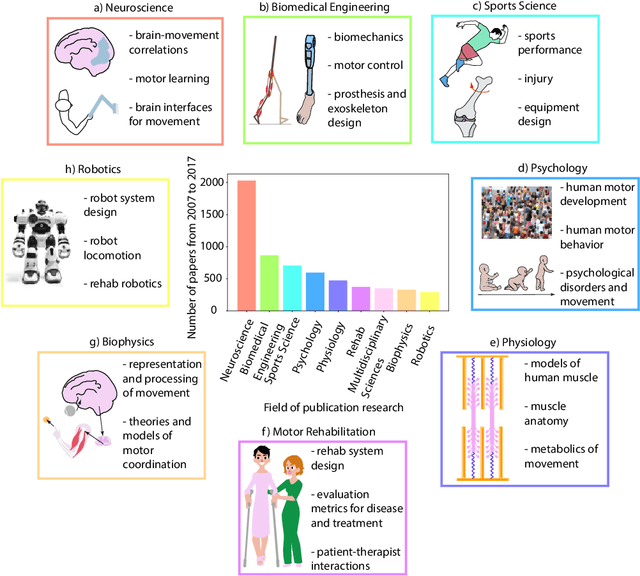

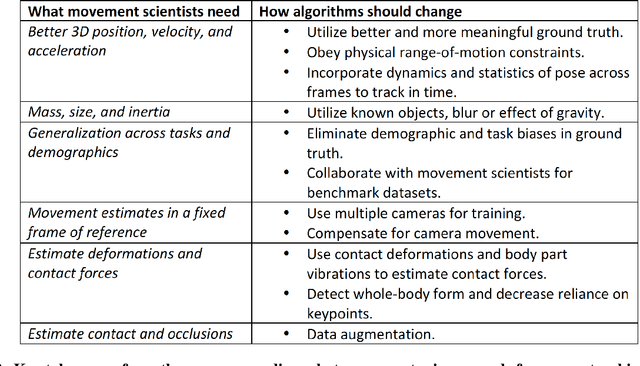
Abstract:Over the last decade, computer science has made progress towards extracting body pose from single camera photographs or videos. This promises to enable movement science to detect disease, quantify movement performance, and take the science out of the lab into the real world. However, current pose tracking algorithms fall short of the needs of movement science; the types of movement data that matter are poorly estimated. For instance, the metrics currently used for evaluating pose tracking algorithms use noisy hand-labeled ground truth data and do not prioritize precision of relevant variables like three-dimensional position, velocity, acceleration, and forces which are crucial for movement science. Here, we introduce the scientific disciplines that use movement data, the types of data they need, and discuss the changes needed to make pose tracking truly transformative for movement science.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge