Sean I. Young
Fast Multi-Stack Slice-to-Volume Reconstruction via Multi-Scale Unrolled Optimization
Jan 12, 2026Abstract:Fully convolutional networks have become the backbone of modern medical imaging due to their ability to learn multi-scale representations and perform end-to-end inference. Yet their potential for slice-to-volume reconstruction (SVR), the task of jointly estimating 3D anatomy and slice poses from misaligned 2D acquisitions, remains underexplored. We introduce a fast convolutional framework that fuses multiple orthogonal 2D slice stacks to recover coherent 3D structure and refines slice alignment through lightweight model-based optimization. Applied to fetal brain MRI, our approach reconstructs high-quality 3D volumes in under 10s, with 1s slice registration and accuracy on par with state-of-the-art iterative SVR pipelines, offering more than speedup. The framework uses non-rigid displacement fields to represent transformations, generalizing to other SVR problems like fetal body and placental MRI. Additionally, the fast inference time paves the way for real-time, scanner-side volumetric feedback during MRI acquisition.
Radio: Rate-Distortion Optimization for Large Language Model Compression
May 05, 2025Abstract:In recent years, the compression of large language models (LLMs) has emerged as a key problem in facilitating LLM deployment on resource-limited devices, reducing compute costs, and mitigating the environmental footprint due to large-scale AI infrastructure. Here, we establish the foundations of LLM quantization from a rate-distortion theory perspective and propose a quantization technique based on simple rate-distortion optimization. Our technique scales to models containing hundreds of billions of weight parameters and offers users the flexibility to compress models, post-training, to a model size or accuracy specified by the user.
Reference-Free 3D Reconstruction of Brain Dissection Photographs with Machine Learning
Mar 13, 2025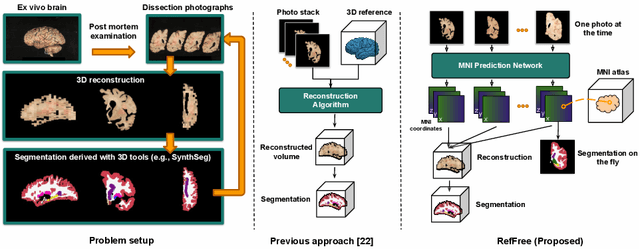
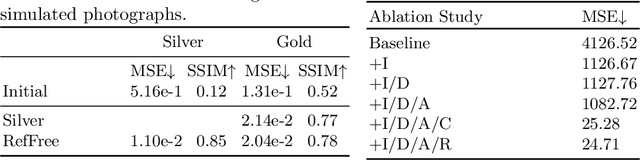
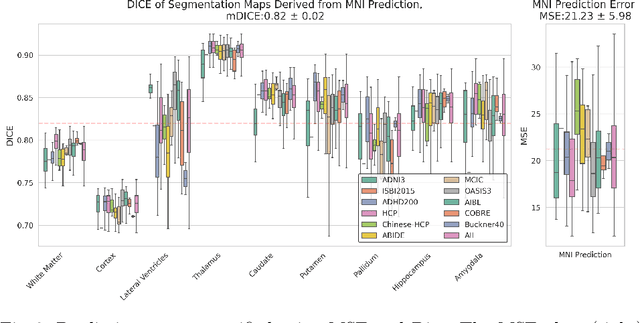
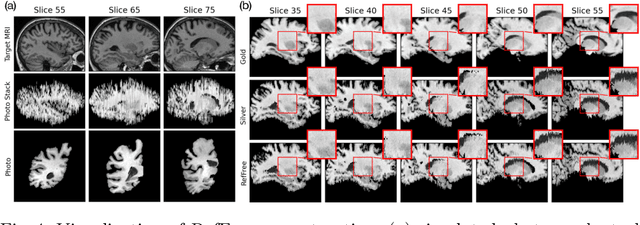
Abstract:Correlation of neuropathology with MRI has the potential to transfer microscopic signatures of pathology to invivo scans. Recently, a classical registration method has been proposed, to build these correlations from 3D reconstructed stacks of dissection photographs, which are routinely taken at brain banks. These photographs bypass the need for exvivo MRI, which is not widely accessible. However, this method requires a full stack of brain slabs and a reference mask (e.g., acquired with a surface scanner), which severely limits the applicability of the technique. Here we propose RefFree, a dissection photograph reconstruction method without external reference. RefFree is a learning approach that estimates the 3D coordinates in the atlas space for every pixel in every photograph; simple least-squares fitting can then be used to compute the 3D reconstruction. As a by-product, RefFree also produces an atlas-based segmentation of the reconstructed stack. RefFree is trained on synthetic photographs generated from digitally sliced 3D MRI data, with randomized appearance for enhanced generalization ability. Experiments on simulated and real data show that RefFree achieves performance comparable to the baseline method without an explicit reference while also enabling reconstruction of partial stacks. Our code is available at https://github.com/lintian-a/reffree.
Foundations of Large Language Model Compression -- Part 1: Weight Quantization
Sep 03, 2024Abstract:In recent years, compression of large language models (LLMs) has emerged as an important problem to allow language model deployment on resource-constrained devices, reduce computational costs, and mitigate the environmental footprint of large-scale AI infrastructure. In this paper, we present the foundations of LLM quantization from a convex optimization perspective and propose a quantization method that builds on these foundations and outperforms previous methods. Our quantization framework, CVXQ, scales to models containing hundreds of billions of weight parameters and provides users with the flexibility to compress models to any specified model size, post-training. A reference implementation of CVXQ can be obtained from https://github.com/seannz/cvxq.
Shape-aware Segmentation of the Placenta in BOLD Fetal MRI Time Series
Dec 08, 2023


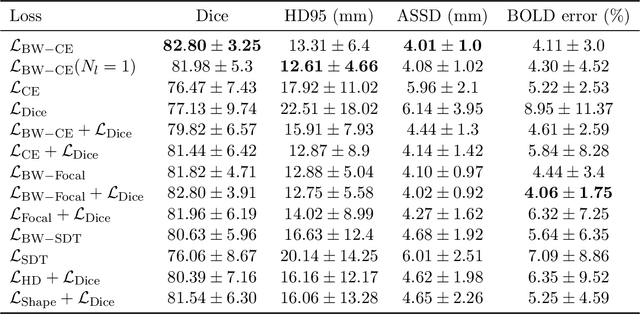
Abstract:Blood oxygen level dependent (BOLD) MRI time series with maternal hyperoxia can assess placental oxygenation and function. Measuring precise BOLD changes in the placenta requires accurate temporal placental segmentation and is confounded by fetal and maternal motion, contractions, and hyperoxia-induced intensity changes. Current BOLD placenta segmentation methods warp a manually annotated subject-specific template to the entire time series. However, as the placenta is a thin, elongated, and highly non-rigid organ subject to large deformations and obfuscated edges, existing work cannot accurately segment the placental shape, especially near boundaries. In this work, we propose a machine learning segmentation framework for placental BOLD MRI and apply it to segmenting each volume in a time series. We use a placental-boundary weighted loss formulation and perform a comprehensive evaluation across several popular segmentation objectives. Our model is trained and tested on a cohort of 91 subjects containing healthy fetuses, fetuses with fetal growth restriction, and mothers with high BMI. Biomedically, our model performs reliably in segmenting volumes in both normoxic and hyperoxic points in the BOLD time series. We further find that boundary-weighting increases placental segmentation performance by 8.3% and 6.0% Dice coefficient for the cross-entropy and signed distance transform objectives, respectively. Our code and trained model is available at https://github.com/mabulnaga/automatic-placenta-segmentation.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2023:017. arXiv admin note: substantial text overlap with arXiv:2208.02895
Quantifying white matter hyperintensity and brain volumes in heterogeneous clinical and low-field portable MRI
Dec 08, 2023
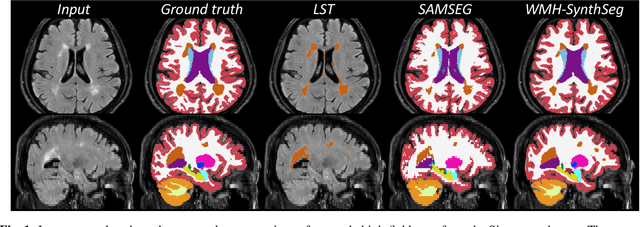
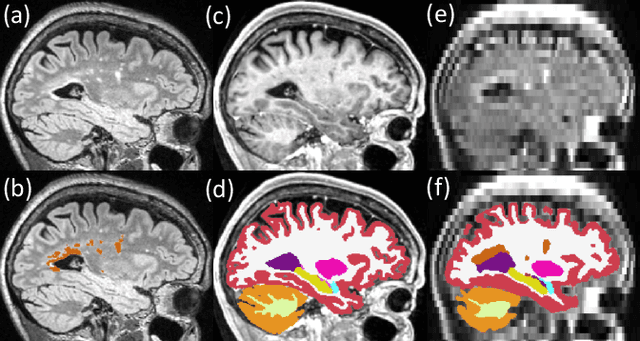

Abstract:Brain atrophy and white matter hyperintensity (WMH) are critical neuroimaging features for ascertaining brain injury in cerebrovascular disease and multiple sclerosis. Automated segmentation and quantification is desirable but existing methods require high-resolution MRI with good signal-to-noise ratio (SNR). This precludes application to clinical and low-field portable MRI (pMRI) scans, thus hampering large-scale tracking of atrophy and WMH progression, especially in underserved areas where pMRI has huge potential. Here we present a method that segments white matter hyperintensity and 36 brain regions from scans of any resolution and contrast (including pMRI) without retraining. We show results on six public datasets and on a private dataset with paired high- and low-field scans (3T and 64mT), where we attain strong correlation between the WMH ($\rho$=.85) and hippocampal volumes (r=.89) estimated at both fields. Our method is publicly available as part of FreeSurfer, at: http://surfer.nmr.mgh.harvard.edu/fswiki/WMH-SynthSeg.
Fully Convolutional Slice-to-Volume Reconstruction for Single-Stack MRI
Dec 05, 2023



Abstract:In magnetic resonance imaging (MRI), slice-to-volume reconstruction (SVR) refers to computational reconstruction of an unknown 3D magnetic resonance volume from stacks of 2D slices corrupted by motion. While promising, current SVR methods require multiple slice stacks for accurate 3D reconstruction, leading to long scans and limiting their use in time-sensitive applications such as fetal fMRI. Here, we propose a SVR method that overcomes the shortcomings of previous work and produces state-of-the-art reconstructions in the presence of extreme inter-slice motion. Inspired by the recent success of single-view depth estimation methods, we formulate SVR as a single-stack motion estimation task and train a fully convolutional network to predict a motion stack for a given slice stack, producing a 3D reconstruction as a byproduct of the predicted motion. Extensive experiments on the SVR of adult and fetal brains demonstrate that our fully convolutional method is twice as accurate as previous SVR methods. Our code is available at github.com/seannz/svr.
A Framework for Interpretability in Machine Learning for Medical Imaging
Oct 02, 2023

Abstract:Interpretability for machine learning models in medical imaging (MLMI) is an important direction of research. However, there is a general sense of murkiness in what interpretability means. Why does the need for interpretability in MLMI arise? What goals does one actually seek to address when interpretability is needed? To answer these questions, we identify a need to formalize the goals and elements of interpretability in MLMI. By reasoning about real-world tasks and goals common in both medical image analysis and its intersection with machine learning, we identify four core elements of interpretability: localization, visual recognizability, physical attribution, and transparency. Overall, this paper formalizes interpretability needs in the context of medical imaging, and our applied perspective clarifies concrete MLMI-specific goals and considerations in order to guide method design and improve real-world usage. Our goal is to provide practical and didactic information for model designers and practitioners, inspire developers of models in the medical imaging field to reason more deeply about what interpretability is achieving, and suggest future directions of interpretability research.
Diffeomorphic Multi-Resolution Deep Learning Registration for Applications in Breast MRI
Sep 24, 2023



Abstract:In breast surgical planning, accurate registration of MR images across patient positions has the potential to improve the localisation of tumours during breast cancer treatment. While learning-based registration methods have recently become the state-of-the-art approach for most medical image registration tasks, these methods have yet to make inroads into breast image registration due to certain difficulties-the lack of rich texture information in breast MR images and the need for the deformations to be diffeomophic. In this work, we propose learning strategies for breast MR image registration that are amenable to diffeomorphic constraints, together with early experimental results from in-silico and in-vivo experiments. One key contribution of this work is a registration network which produces superior registration outcomes for breast images in addition to providing diffeomorphic guarantees.
SUD$^2$: Supervision by Denoising Diffusion Models for Image Reconstruction
Apr 03, 2023



Abstract:Many imaging inverse problems$\unicode{x2014}$such as image-dependent in-painting and dehazing$\unicode{x2014}$are challenging because their forward models are unknown or depend on unknown latent parameters. While one can solve such problems by training a neural network with vast quantities of paired training data, such paired training data is often unavailable. In this paper, we propose a generalized framework for training image reconstruction networks when paired training data is scarce. In particular, we demonstrate the ability of image denoising algorithms and, by extension, denoising diffusion models to supervise network training in the absence of paired training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge