Clinton J. Wang
Fast Multi-Stack Slice-to-Volume Reconstruction via Multi-Scale Unrolled Optimization
Jan 12, 2026Abstract:Fully convolutional networks have become the backbone of modern medical imaging due to their ability to learn multi-scale representations and perform end-to-end inference. Yet their potential for slice-to-volume reconstruction (SVR), the task of jointly estimating 3D anatomy and slice poses from misaligned 2D acquisitions, remains underexplored. We introduce a fast convolutional framework that fuses multiple orthogonal 2D slice stacks to recover coherent 3D structure and refines slice alignment through lightweight model-based optimization. Applied to fetal brain MRI, our approach reconstructs high-quality 3D volumes in under 10s, with 1s slice registration and accuracy on par with state-of-the-art iterative SVR pipelines, offering more than speedup. The framework uses non-rigid displacement fields to represent transformations, generalizing to other SVR problems like fetal body and placental MRI. Additionally, the fast inference time paves the way for real-time, scanner-side volumetric feedback during MRI acquisition.
EnigmaEval: A Benchmark of Long Multimodal Reasoning Challenges
Feb 13, 2025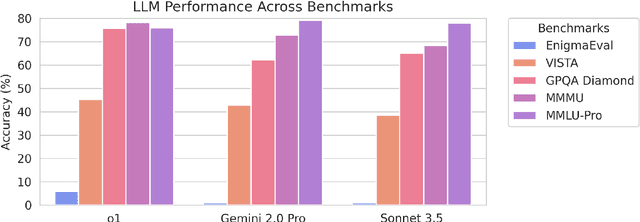

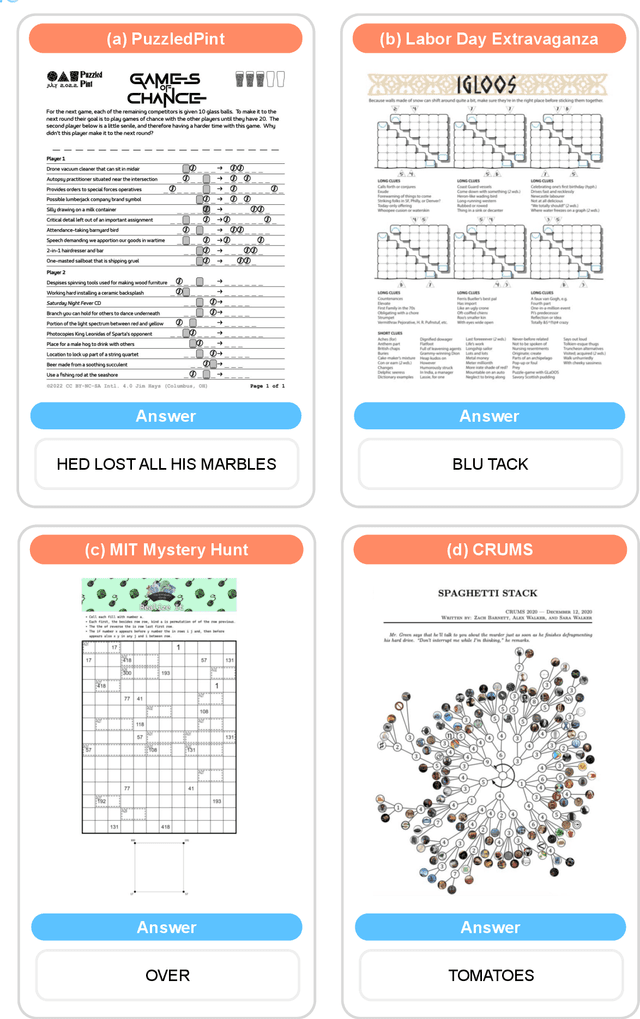

Abstract:As language models master existing reasoning benchmarks, we need new challenges to evaluate their cognitive frontiers. Puzzle-solving events are rich repositories of challenging multimodal problems that test a wide range of advanced reasoning and knowledge capabilities, making them a unique testbed for evaluating frontier language models. We introduce EnigmaEval, a dataset of problems and solutions derived from puzzle competitions and events that probes models' ability to perform implicit knowledge synthesis and multi-step deductive reasoning. Unlike existing reasoning and knowledge benchmarks, puzzle solving challenges models to discover hidden connections between seemingly unrelated pieces of information to uncover solution paths. The benchmark comprises 1184 puzzles of varying complexity -- each typically requiring teams of skilled solvers hours to days to complete -- with unambiguous, verifiable solutions that enable efficient evaluation. State-of-the-art language models achieve extremely low accuracy on these puzzles, even lower than other difficult benchmarks such as Humanity's Last Exam, unveiling models' shortcomings when challenged with problems requiring unstructured and lateral reasoning.
Learning General-Purpose Biomedical Volume Representations using Randomized Synthesis
Nov 04, 2024Abstract:Current volumetric biomedical foundation models struggle to generalize as public 3D datasets are small and do not cover the broad diversity of medical procedures, conditions, anatomical regions, and imaging protocols. We address this by creating a representation learning method that instead anticipates strong domain shifts at training time itself. We first propose a data engine that synthesizes highly variable training samples that enable generalization to new biomedical contexts. To then train a single 3D network for any voxel-level task, we develop a contrastive learning method that pretrains the network to be stable against nuisance imaging variation simulated by the data engine, a key inductive bias for generalization. This network's features can be used as robust representations of input images for downstream tasks and its weights provide a strong, dataset-agnostic initialization for finetuning on new datasets. As a result, we set new standards across both multimodality registration and few-shot segmentation, a first for any 3D biomedical vision model, all without (pre-)training on any existing dataset of real images.
Shape-aware Segmentation of the Placenta in BOLD Fetal MRI Time Series
Dec 08, 2023


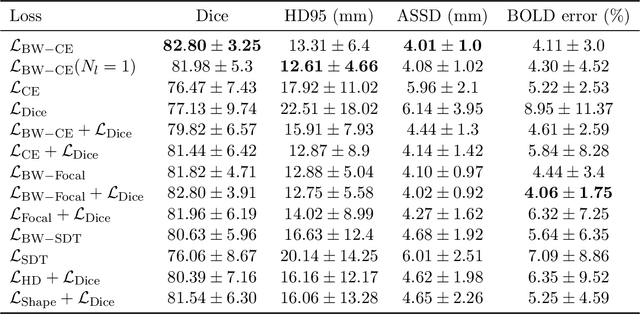
Abstract:Blood oxygen level dependent (BOLD) MRI time series with maternal hyperoxia can assess placental oxygenation and function. Measuring precise BOLD changes in the placenta requires accurate temporal placental segmentation and is confounded by fetal and maternal motion, contractions, and hyperoxia-induced intensity changes. Current BOLD placenta segmentation methods warp a manually annotated subject-specific template to the entire time series. However, as the placenta is a thin, elongated, and highly non-rigid organ subject to large deformations and obfuscated edges, existing work cannot accurately segment the placental shape, especially near boundaries. In this work, we propose a machine learning segmentation framework for placental BOLD MRI and apply it to segmenting each volume in a time series. We use a placental-boundary weighted loss formulation and perform a comprehensive evaluation across several popular segmentation objectives. Our model is trained and tested on a cohort of 91 subjects containing healthy fetuses, fetuses with fetal growth restriction, and mothers with high BMI. Biomedically, our model performs reliably in segmenting volumes in both normoxic and hyperoxic points in the BOLD time series. We further find that boundary-weighting increases placental segmentation performance by 8.3% and 6.0% Dice coefficient for the cross-entropy and signed distance transform objectives, respectively. Our code and trained model is available at https://github.com/mabulnaga/automatic-placenta-segmentation.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2023:017. arXiv admin note: substantial text overlap with arXiv:2208.02895
Dynamic Neural Fields for Learning Atlases of 4D Fetal MRI Time-series
Nov 06, 2023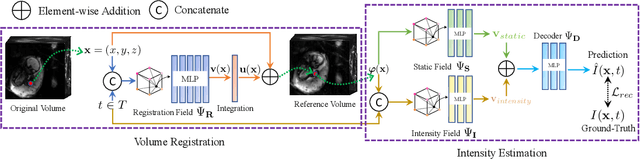
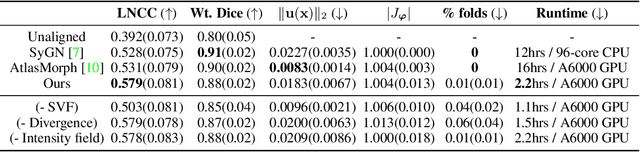
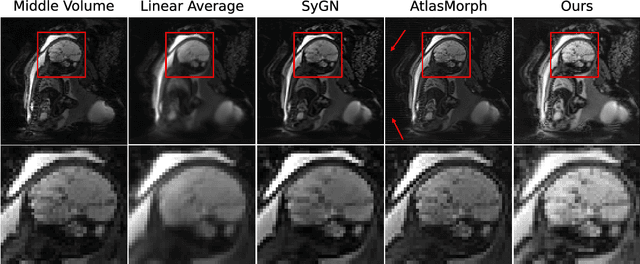
Abstract:We present a method for fast biomedical image atlas construction using neural fields. Atlases are key to biomedical image analysis tasks, yet conventional and deep network estimation methods remain time-intensive. In this preliminary work, we frame subject-specific atlas building as learning a neural field of deformable spatiotemporal observations. We apply our method to learning subject-specific atlases and motion stabilization of dynamic BOLD MRI time-series of fetuses in utero. Our method yields high-quality atlases of fetal BOLD time-series with $\sim$5-7$\times$ faster convergence compared to existing work. While our method slightly underperforms well-tuned baselines in terms of anatomical overlap, it estimates templates significantly faster, thus enabling rapid processing and stabilization of large databases of 4D dynamic MRI acquisitions. Code is available at https://github.com/Kidrauh/neural-atlasing
Interpolating between Images with Diffusion Models
Jul 24, 2023



Abstract:One little-explored frontier of image generation and editing is the task of interpolating between two input images, a feature missing from all currently deployed image generation pipelines. We argue that such a feature can expand the creative applications of such models, and propose a method for zero-shot interpolation using latent diffusion models. We apply interpolation in the latent space at a sequence of decreasing noise levels, then perform denoising conditioned on interpolated text embeddings derived from textual inversion and (optionally) subject poses. For greater consistency, or to specify additional criteria, we can generate several candidates and use CLIP to select the highest quality image. We obtain convincing interpolations across diverse subject poses, image styles, and image content, and show that standard quantitative metrics such as FID are insufficient to measure the quality of an interpolation. Code and data are available at https://clintonjwang.github.io/interpolation.
Automatic Segmentation of the Placenta in BOLD MRI Time Series
Aug 04, 2022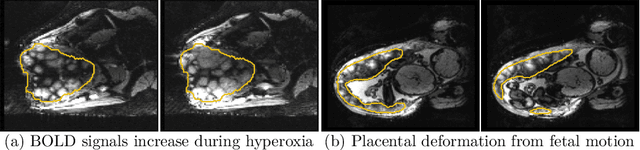
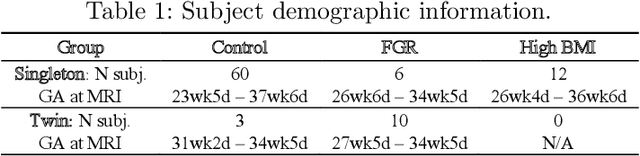
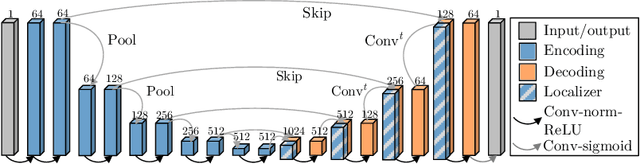

Abstract:Blood oxygen level dependent (BOLD) MRI with maternal hyperoxia can assess oxygen transport within the placenta and has emerged as a promising tool to study placental function. Measuring signal changes over time requires segmenting the placenta in each volume of the time series. Due to the large number of volumes in the BOLD time series, existing studies rely on registration to map all volumes to a manually segmented template. As the placenta can undergo large deformation due to fetal motion, maternal motion, and contractions, this approach often results in a large number of discarded volumes, where the registration approach fails. In this work, we propose a machine learning model based on a U-Net neural network architecture to automatically segment the placenta in BOLD MRI and apply it to segmenting each volume in a time series. We use a boundary-weighted loss function to accurately capture the placental shape. Our model is trained and tested on a cohort of 91 subjects containing healthy fetuses, fetuses with fetal growth restriction, and mothers with high BMI. We achieve a Dice score of 0.83+/-0.04 when matching with ground truth labels and our model performs reliably in segmenting volumes in both normoxic and hyperoxic points in the BOLD time series. Our code and trained model are available at https://github.com/mabulnaga/automatic-placenta-segmentation.
Deep Learning on Implicit Neural Datasets
Jun 02, 2022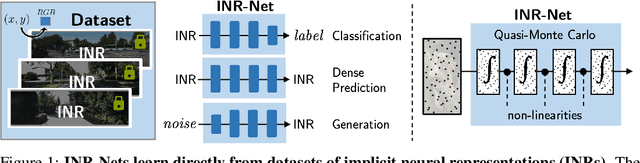
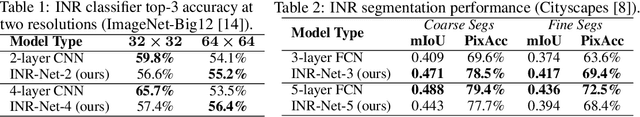
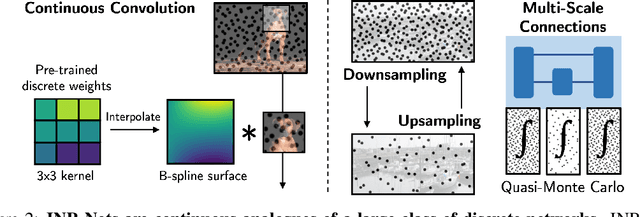
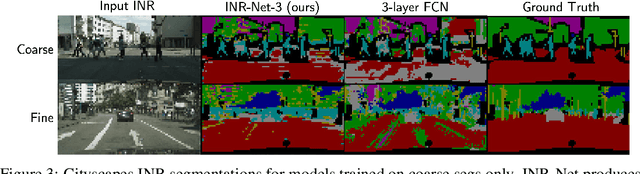
Abstract:Implicit neural representations (INRs) have become fast, lightweight tools for storing continuous data, but to date there is no general method for learning directly with INRs as a data representation. We introduce a principled deep learning framework for learning and inference directly with INRs of any type without reverting to grid-based features or operations. Our INR-Nets evaluate INRs on a low discrepancy sequence, enabling quasi-Monte Carlo (QMC) integration throughout the network. We prove INR-Nets are universal approximators on a large class of maps between $L^2$ functions. Additionally, INR-Nets have convergent gradients under the empirical measure, enabling backpropagation. We design INR-Nets as a continuous generalization of discrete networks, enabling them to be initialized with pre-trained models. We demonstrate learning of INR-Nets on classification (INR$\to$label) and segmentation (INR$\to$INR) tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge