Neel Dey
PolyPose: Localizing Deformable Anatomy in 3D from Sparse 2D X-ray Images using Polyrigid Transforms
May 25, 2025Abstract:Determining the 3D pose of a patient from a limited set of 2D X-ray images is a critical task in interventional settings. While preoperative volumetric imaging (e.g., CT and MRI) provides precise 3D localization and visualization of anatomical targets, these modalities cannot be acquired during procedures, where fast 2D imaging (X-ray) is used instead. To integrate volumetric guidance into intraoperative procedures, we present PolyPose, a simple and robust method for deformable 2D/3D registration. PolyPose parameterizes complex 3D deformation fields as a composition of rigid transforms, leveraging the biological constraint that individual bones do not bend in typical motion. Unlike existing methods that either assume no inter-joint movement or fail outright in this under-determined setting, our polyrigid formulation enforces anatomically plausible priors that respect the piecewise rigid nature of human movement. This approach eliminates the need for expensive deformation regularizers that require patient- and procedure-specific hyperparameter optimization. Across extensive experiments on diverse datasets from orthopedic surgery and radiotherapy, we show that this strong inductive bias enables PolyPose to successfully align the patient's preoperative volume to as few as two X-ray images, thereby providing crucial 3D guidance in challenging sparse-view and limited-angle settings where current registration methods fail.
MultiMorph: On-demand Atlas Construction
Mar 31, 2025Abstract:We present MultiMorph, a fast and efficient method for constructing anatomical atlases on the fly. Atlases capture the canonical structure of a collection of images and are essential for quantifying anatomical variability across populations. However, current atlas construction methods often require days to weeks of computation, thereby discouraging rapid experimentation. As a result, many scientific studies rely on suboptimal, precomputed atlases from mismatched populations, negatively impacting downstream analyses. MultiMorph addresses these challenges with a feedforward model that rapidly produces high-quality, population-specific atlases in a single forward pass for any 3D brain dataset, without any fine-tuning or optimization. MultiMorph is based on a linear group-interaction layer that aggregates and shares features within the group of input images. Further, by leveraging auxiliary synthetic data, MultiMorph generalizes to new imaging modalities and population groups at test-time. Experimentally, MultiMorph outperforms state-of-the-art optimization-based and learning-based atlas construction methods in both small and large population settings, with a 100-fold reduction in time. This makes MultiMorph an accessible framework for biomedical researchers without machine learning expertise, enabling rapid, high-quality atlas generation for diverse studies.
Rapid patient-specific neural networks for intraoperative X-ray to volume registration
Mar 20, 2025Abstract:The integration of artificial intelligence in image-guided interventions holds transformative potential, promising to extract 3D geometric and quantitative information from conventional 2D imaging modalities during complex procedures. Achieving this requires the rapid and precise alignment of 2D intraoperative images (e.g., X-ray) with 3D preoperative volumes (e.g., CT, MRI). However, current 2D/3D registration methods fail across the broad spectrum of procedures dependent on X-ray guidance: traditional optimization techniques require custom parameter tuning for each subject, whereas neural networks trained on small datasets do not generalize to new patients or require labor-intensive manual annotations, increasing clinical burden and precluding application to new anatomical targets. To address these challenges, we present xvr, a fully automated framework for training patient-specific neural networks for 2D/3D registration. xvr uses physics-based simulation to generate abundant high-quality training data from a patient's own preoperative volumetric imaging, thereby overcoming the inherently limited ability of supervised models to generalize to new patients and procedures. Furthermore, xvr requires only 5 minutes of training per patient, making it suitable for emergency interventions as well as planned procedures. We perform the largest evaluation of a 2D/3D registration algorithm on real X-ray data to date and find that xvr robustly generalizes across a diverse dataset comprising multiple anatomical structures, imaging modalities, and hospitals. Across surgical tasks, xvr achieves submillimeter-accurate registration at intraoperative speeds, improving upon existing methods by an order of magnitude. xvr is released as open-source software freely available at https://github.com/eigenvivek/xvr.
Synthesizing Individualized Aging Brains in Health and Disease with Generative Models and Parallel Transport
Feb 28, 2025



Abstract:Simulating prospective magnetic resonance imaging (MRI) scans from a given individual brain image is challenging, as it requires accounting for canonical changes in aging and/or disease progression while also considering the individual brain's current status and unique characteristics. While current deep generative models can produce high-resolution anatomically accurate templates for population-wide studies, their ability to predict future aging trajectories for individuals remains limited, particularly in capturing subject-specific neuroanatomical variations over time. In this study, we introduce Individualized Brain Synthesis (InBrainSyn), a framework for synthesizing high-resolution subject-specific longitudinal MRI scans that simulate neurodegeneration in both Alzheimer's disease (AD) and normal aging. InBrainSyn uses a parallel transport algorithm to adapt the population-level aging trajectories learned by a generative deep template network, enabling individualized aging synthesis. As InBrainSyn uses diffeomorphic transformations to simulate aging, the synthesized images are topologically consistent with the original anatomy by design. We evaluated InBrainSyn both quantitatively and qualitatively on AD and healthy control cohorts from the Open Access Series of Imaging Studies - version 3 dataset. Experimentally, InBrainSyn can also model neuroanatomical transitions between normal aging and AD. An evaluation of an external set supports its generalizability. Overall, with only a single baseline scan, InBrainSyn synthesizes realistic 3D spatiotemporal T1w MRI scans, producing personalized longitudinal aging trajectories. The code for InBrainSyn is available at: https://github.com/Fjr9516/InBrainSyn.
Differentiable Voxel-based X-ray Rendering Improves Sparse-View 3D CBCT Reconstruction
Dec 02, 2024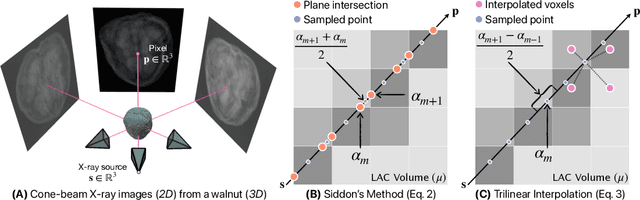
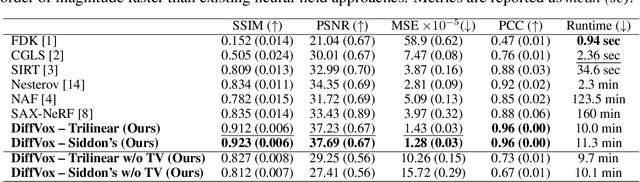
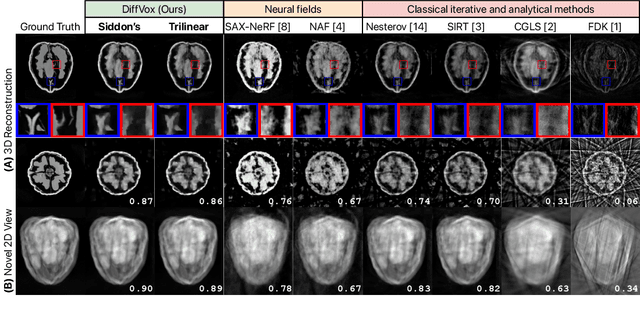
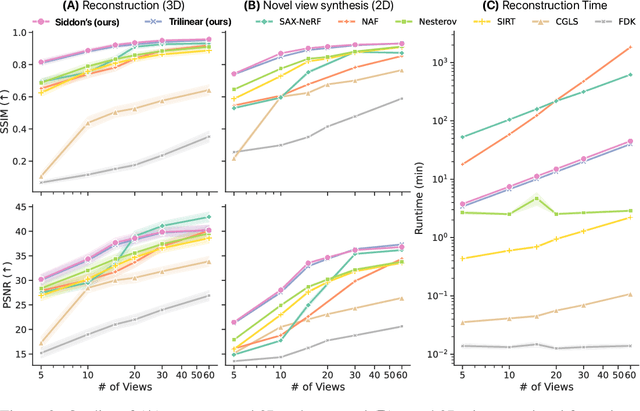
Abstract:We present DiffVox, a self-supervised framework for Cone-Beam Computed Tomography (CBCT) reconstruction by directly optimizing a voxelgrid representation using physics-based differentiable X-ray rendering. Further, we investigate how the different implementations of the X-ray image formation model in the renderer affect the quality of 3D reconstruction and novel view synthesis. When combined with our regularized voxel-based learning framework, we find that using an exact implementation of the discrete Beer-Lambert law for X-ray attenuation in the renderer outperforms both widely used iterative CBCT reconstruction algorithms and modern neural field approaches, particularly when given only a few input views. As a result, we reconstruct high-fidelity 3D CBCT volumes from fewer X-rays, potentially reducing ionizing radiation exposure and improving diagnostic utility. Our implementation is available at https://github.com/hossein-momeni/DiffVox.
Equivariant spatio-hemispherical networks for diffusion MRI deconvolution
Nov 18, 2024



Abstract:Each voxel in a diffusion MRI (dMRI) image contains a spherical signal corresponding to the direction and strength of water diffusion in the brain. This paper advances the analysis of such spatio-spherical data by developing convolutional network layers that are equivariant to the $\mathbf{E(3) \times SO(3)}$ group and account for the physical symmetries of dMRI including rotations, translations, and reflections of space alongside voxel-wise rotations. Further, neuronal fibers are typically antipodally symmetric, a fact we leverage to construct highly efficient spatio-hemispherical graph convolutions to accelerate the analysis of high-dimensional dMRI data. In the context of sparse spherical fiber deconvolution to recover white matter microstructure, our proposed equivariant network layers yield substantial performance and efficiency gains, leading to better and more practical resolution of crossing neuronal fibers and fiber tractography. These gains are experimentally consistent across both simulation and in vivo human datasets.
Learning General-Purpose Biomedical Volume Representations using Randomized Synthesis
Nov 04, 2024Abstract:Current volumetric biomedical foundation models struggle to generalize as public 3D datasets are small and do not cover the broad diversity of medical procedures, conditions, anatomical regions, and imaging protocols. We address this by creating a representation learning method that instead anticipates strong domain shifts at training time itself. We first propose a data engine that synthesizes highly variable training samples that enable generalization to new biomedical contexts. To then train a single 3D network for any voxel-level task, we develop a contrastive learning method that pretrains the network to be stable against nuisance imaging variation simulated by the data engine, a key inductive bias for generalization. This network's features can be used as robust representations of input images for downstream tasks and its weights provide a strong, dataset-agnostic initialization for finetuning on new datasets. As a result, we set new standards across both multimodality registration and few-shot segmentation, a first for any 3D biomedical vision model, all without (pre-)training on any existing dataset of real images.
Geo-UNet: A Geometrically Constrained Neural Framework for Clinical-Grade Lumen Segmentation in Intravascular Ultrasound
Aug 09, 2024



Abstract:Precisely estimating lumen boundaries in intravascular ultrasound (IVUS) is needed for sizing interventional stents to treat deep vein thrombosis (DVT). Unfortunately, current segmentation networks like the UNet lack the precision needed for clinical adoption in IVUS workflows. This arises due to the difficulty of automatically learning accurate lumen contour from limited training data while accounting for the radial geometry of IVUS imaging. We propose the Geo-UNet framework to address these issues via a design informed by the geometry of the lumen contour segmentation task. We first convert the input data and segmentation targets from Cartesian to polar coordinates. Starting from a convUNet feature extractor, we propose a two-task setup, one for conventional pixel-wise labeling and the other for single boundary lumen-contour localization. We directly combine the two predictions by passing the predicted lumen contour through a new activation (named CDFeLU) to filter out spurious pixel-wise predictions. Our unified loss function carefully balances area-based, distance-based, and contour-based penalties to provide near clinical-grade generalization in unseen patient data. We also introduce a lightweight, inference-time technique to enhance segmentation smoothness. The efficacy of our framework on a venous IVUS dataset is shown against state-of-the-art models.
SE-Equivariant and Noise-Invariant 3D Motion Tracking in Medical Images
Dec 21, 2023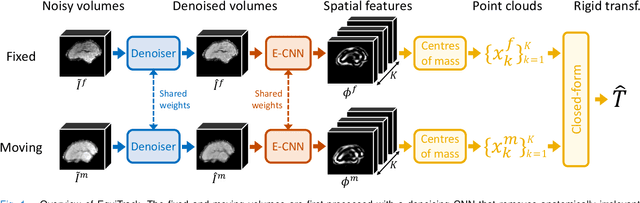
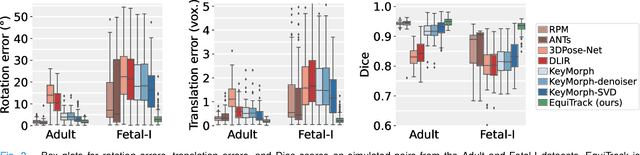
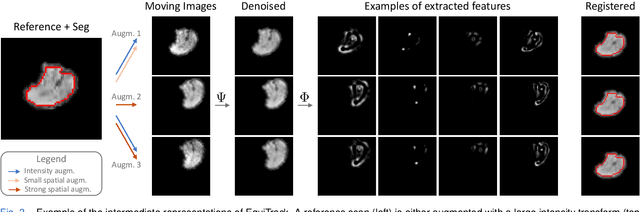
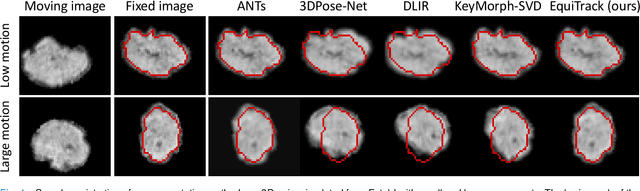
Abstract:Rigid motion tracking is paramount in many medical imaging applications where movements need to be detected, corrected, or accounted for. Modern strategies rely on convolutional neural networks (CNN) and pose this problem as rigid registration. Yet, CNNs do not exploit natural symmetries in this task, as they are equivariant to translations (their outputs shift with their inputs) but not to rotations. Here we propose EquiTrack, the first method that uses recent steerable SE(3)-equivariant CNNs (E-CNN) for motion tracking. While steerable E-CNNs can extract corresponding features across different poses, testing them on noisy medical images reveals that they do not have enough learning capacity to learn noise invariance. Thus, we introduce a hybrid architecture that pairs a denoiser with an E-CNN to decouple the processing of anatomically irrelevant intensity features from the extraction of equivariant spatial features. Rigid transforms are then estimated in closed-form. EquiTrack outperforms state-of-the-art learning and optimisation methods for motion tracking in adult brain MRI and fetal MRI time series. Our code is available at github.com/BBillot/equitrack.
Intraoperative 2D/3D Image Registration via Differentiable X-ray Rendering
Dec 11, 2023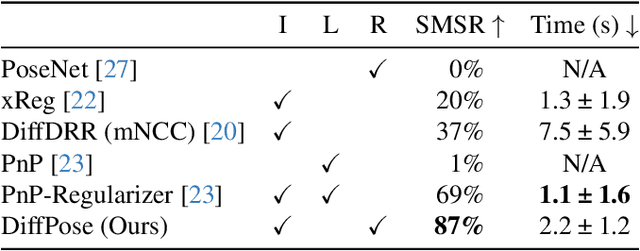
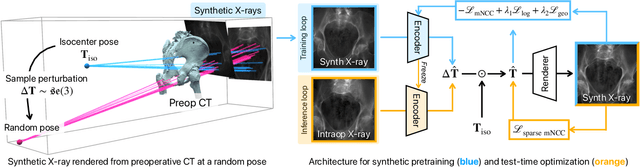
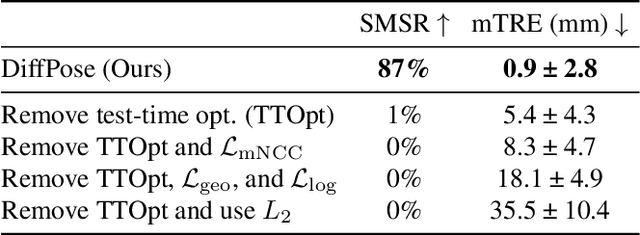
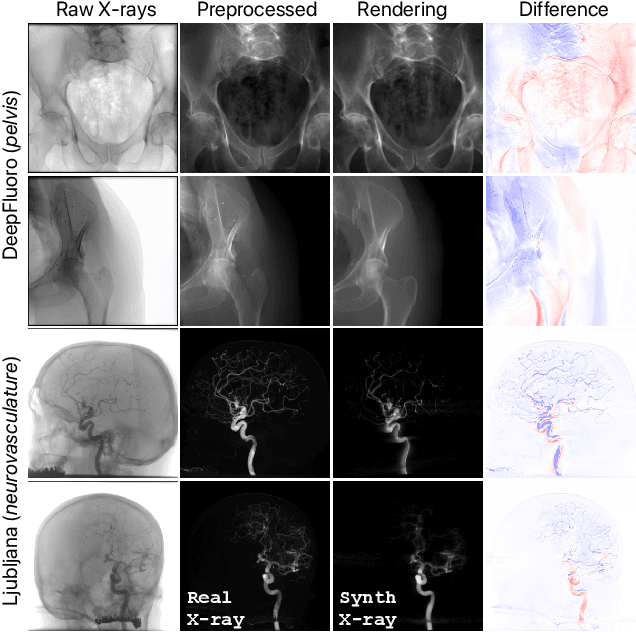
Abstract:Surgical decisions are informed by aligning rapid portable 2D intraoperative images (e.g., X-rays) to a high-fidelity 3D preoperative reference scan (e.g., CT). 2D/3D image registration often fails in practice: conventional optimization methods are prohibitively slow and susceptible to local minima, while neural networks trained on small datasets fail on new patients or require impractical landmark supervision. We present DiffPose, a self-supervised approach that leverages patient-specific simulation and differentiable physics-based rendering to achieve accurate 2D/3D registration without relying on manually labeled data. Preoperatively, a CNN is trained to regress the pose of a randomly oriented synthetic X-ray rendered from the preoperative CT. The CNN then initializes rapid intraoperative test-time optimization that uses the differentiable X-ray renderer to refine the solution. Our work further proposes several geometrically principled methods for sampling camera poses from $\mathbf{SE}(3)$, for sparse differentiable rendering, and for driving registration in the tangent space $\mathfrak{se}(3)$ with geodesic and multiscale locality-sensitive losses. DiffPose achieves sub-millimeter accuracy across surgical datasets at intraoperative speeds, improving upon existing unsupervised methods by an order of magnitude and even outperforming supervised baselines. Our code is available at https://github.com/eigenvivek/DiffPose.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge