Malte Hoffmann
Harvard University
Unified Brain Surface and Volume Registration
Dec 22, 2025Abstract:Accurate registration of brain MRI scans is fundamental for cross-subject analysis in neuroscientific studies. This involves aligning both the cortical surface of the brain and the interior volume. Traditional methods treat volumetric and surface-based registration separately, which often leads to inconsistencies that limit downstream analyses. We propose a deep learning framework, NeurAlign, that registers $3$D brain MRI images by jointly aligning both cortical and subcortical regions through a unified volume-and-surface-based representation. Our approach leverages an intermediate spherical coordinate space to bridge anatomical surface topology with volumetric anatomy, enabling consistent and anatomically accurate alignment. By integrating spherical registration into the learning, our method ensures geometric coherence between volume and surface domains. In a series of experiments on both in-domain and out-of-domain datasets, our method consistently outperforms both classical and machine learning-based registration methods -- improving the Dice score by up to 7 points while maintaining regular deformation fields. Additionally, it is orders of magnitude faster than the standard method for this task, and is simpler to use because it requires no additional inputs beyond an MRI scan. With its superior accuracy, fast inference, and ease of use, NeurAlign sets a new standard for joint cortical and subcortical registration.
Deep learning water-unsuppressed MRSI at ultra-high field for simultaneous quantitative metabolic, susceptibility and myelin water imaging
Dec 16, 2025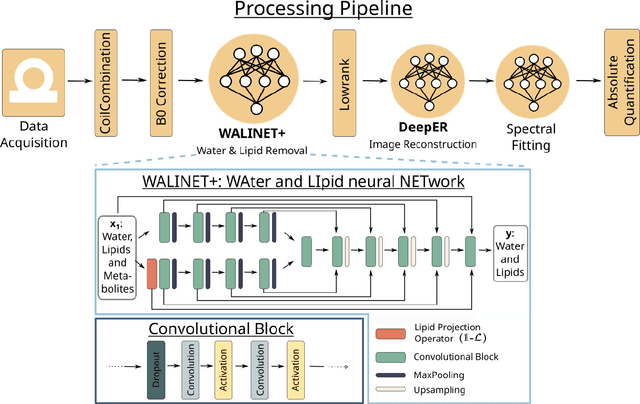
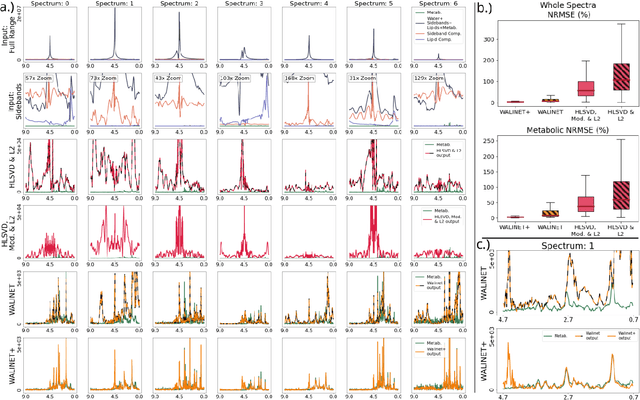
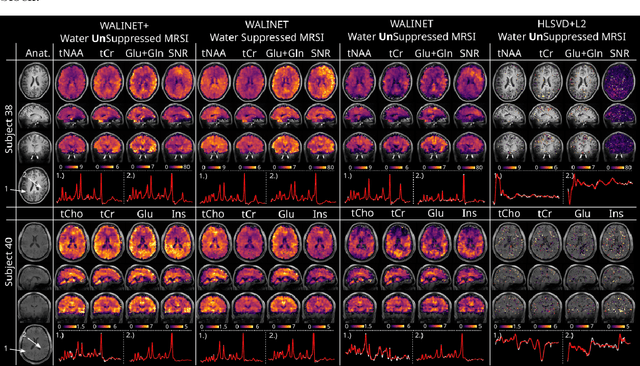
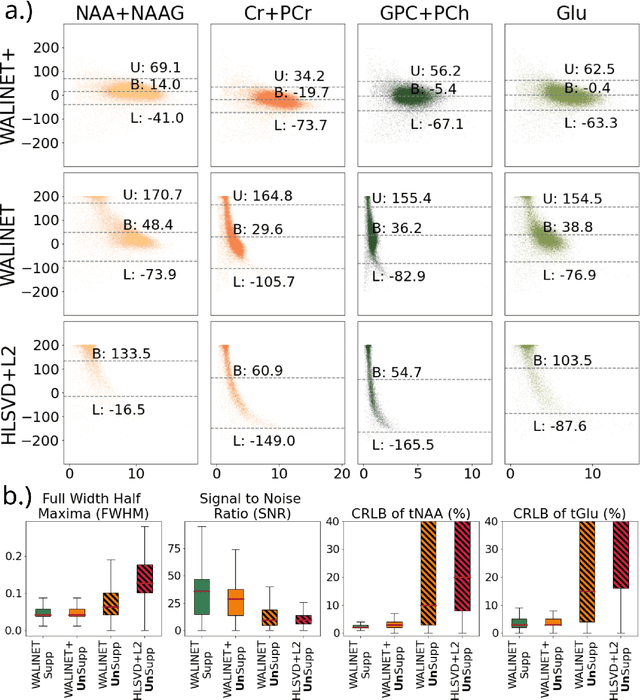
Abstract:Purpose: Magnetic Resonance Spectroscopic Imaging (MRSI) maps endogenous brain metabolism while suppressing the overwhelming water signal. Water-unsuppressed MRSI (wu-MRSI) allows simultaneous imaging of water and metabolites, but large water sidebands cause challenges for metabolic fitting. We developed an end-to-end deep-learning pipeline to overcome these challenges at ultra-high field. Methods:Fast high-resolution wu-MRSI was acquired at 7T with non-cartesian ECCENTRIC sampling and ultra-short echo time. A water and lipid removal network (WALINET+) was developed to remove lipids, water signal, and sidebands. MRSI reconstruction was performed by DeepER and a physics-informed network for metabolite fitting. Water signal was used for absolute metabolite quantification, quantitative susceptibility mapping (QSM), and myelin water fraction imaging (MWF). Results: WALINET+ provided the lowest NRMSE (< 2%) in simulations and in vivo the smallest bias (< 20%) and limits-of-agreement (+-63%) between wu-MRSI and ws-MRSI scans. Several metabolites such as creatine and glutamate showed higher SNR in wu-MRSI. QSM and MWF obtained from wu-MRSI and GRE showed good agreement with 0 ppm/5.5% bias and +-0.05 ppm/ +- 12.75% limits-of-agreement. Conclusion: High-quality metabolic, QSM, and MWF mapping of the human brain can be obtained simultaneously by ECCENTRIC wu-MRSI at 7T with 2 mm isotropic resolution in 12 min. WALINET+ robustly removes water sidebands while preserving metabolite signal, eliminating the need for water suppression and separate water acquisitions.
Domain-randomized deep learning for neuroimage analysis
Jul 17, 2025Abstract:Deep learning has revolutionized neuroimage analysis by delivering unprecedented speed and accuracy. However, the narrow scope of many training datasets constrains model robustness and generalizability. This challenge is particularly acute in magnetic resonance imaging (MRI), where image appearance varies widely across pulse sequences and scanner hardware. A recent domain-randomization strategy addresses the generalization problem by training deep neural networks on synthetic images with randomized intensities and anatomical content. By generating diverse data from anatomical segmentation maps, the approach enables models to accurately process image types unseen during training, without retraining or fine-tuning. It has demonstrated effectiveness across modalities including MRI, computed tomography, positron emission tomography, and optical coherence tomography, as well as beyond neuroimaging in ultrasound, electron and fluorescence microscopy, and X-ray microtomography. This tutorial paper reviews the principles, implementation, and potential of the synthesis-driven training paradigm. It highlights key benefits, such as improved generalization and resistance to overfitting, while discussing trade-offs such as increased computational demands. Finally, the article explores practical considerations for adopting the technique, aiming to accelerate the development of generalizable tools that make deep learning more accessible to domain experts without extensive computational resources or machine learning knowledge.
MindGrab for BrainChop: Fast and Accurate Skull Stripping for Command Line and Browser
Jun 13, 2025Abstract:We developed MindGrab, a parameter- and memory-efficient deep fully-convolutional model for volumetric skull-stripping in head images of any modality. Its architecture, informed by a spectral interpretation of dilated convolutions, was trained exclusively on modality-agnostic synthetic data. MindGrab was evaluated on a retrospective dataset of 606 multimodal adult-brain scans (T1, T2, DWI, MRA, PDw MRI, EPI, CT, PET) sourced from the SynthStrip dataset. Performance was benchmarked against SynthStrip, ROBEX, and BET using Dice scores, with Wilcoxon signed-rank significance tests. MindGrab achieved a mean Dice score of 95.9 with standard deviation (SD) 1.6 across modalities, significantly outperforming classical methods (ROBEX: 89.1 SD 7.7, P < 0.05; BET: 85.2 SD 14.4, P < 0.05). Compared to SynthStrip (96.5 SD 1.1, P=0.0352), MindGrab delivered equivalent or superior performance in nearly half of the tested scenarios, with minor differences (<3% Dice) in the others. MindGrab utilized 95% fewer parameters (146,237 vs. 2,566,561) than SynthStrip. This efficiency yielded at least 2x faster inference, 50% lower memory usage on GPUs, and enabled exceptional performance (e.g., 10-30x speedup, and up to 30x memory reduction) and accessibility on a wider range of hardware, including systems without high-end GPUs. MindGrab delivers state-of-the-art accuracy with dramatically lower resource demands, supported in brainchop-cli (https://pypi.org/project/brainchop/) and at brainchop.org.
MultiMorph: On-demand Atlas Construction
Mar 31, 2025Abstract:We present MultiMorph, a fast and efficient method for constructing anatomical atlases on the fly. Atlases capture the canonical structure of a collection of images and are essential for quantifying anatomical variability across populations. However, current atlas construction methods often require days to weeks of computation, thereby discouraging rapid experimentation. As a result, many scientific studies rely on suboptimal, precomputed atlases from mismatched populations, negatively impacting downstream analyses. MultiMorph addresses these challenges with a feedforward model that rapidly produces high-quality, population-specific atlases in a single forward pass for any 3D brain dataset, without any fine-tuning or optimization. MultiMorph is based on a linear group-interaction layer that aggregates and shares features within the group of input images. Further, by leveraging auxiliary synthetic data, MultiMorph generalizes to new imaging modalities and population groups at test-time. Experimentally, MultiMorph outperforms state-of-the-art optimization-based and learning-based atlas construction methods in both small and large population settings, with a 100-fold reduction in time. This makes MultiMorph an accessible framework for biomedical researchers without machine learning expertise, enabling rapid, high-quality atlas generation for diverse studies.
Search Wide, Focus Deep: Automated Fetal Brain Extraction with Sparse Training Data
Oct 29, 2024



Abstract:Automated fetal brain extraction from full-uterus MRI is a challenging task due to variable head sizes, orientations, complex anatomy, and prevalent artifacts. While deep-learning (DL) models trained on synthetic images have been successful in adult brain extraction, adapting these networks for fetal MRI is difficult due to the sparsity of labeled data, leading to increased false-positive predictions. To address this challenge, we propose a test-time strategy that reduces false positives in networks trained on sparse, synthetic labels. The approach uses a breadth-fine search (BFS) to identify a subvolume likely to contain the fetal brain, followed by a deep-focused sliding window (DFS) search to refine the extraction, pooling predictions to minimize false positives. We train models at different window sizes using synthetic images derived from a small number of fetal brain label maps, augmented with random geometric shapes. Each model is trained on diverse head positions and scales, including cases with partial or no brain tissue. Our framework matches state-of-the-art brain extraction methods on clinical HASTE scans of third-trimester fetuses and exceeds them by up to 5\% in terms of Dice in the second trimester as well as EPI scans across both trimesters. Our results demonstrate the utility of a sliding-window approach and combining predictions from several models trained on synthetic images, for improving brain-extraction accuracy by progressively refining regions of interest and minimizing the risk of missing brain mask slices or misidentifying other tissues as brain.
WALINET: A water and lipid identification convolutional Neural Network for nuisance signal removal in 1H MR Spectroscopic Imaging
Oct 01, 2024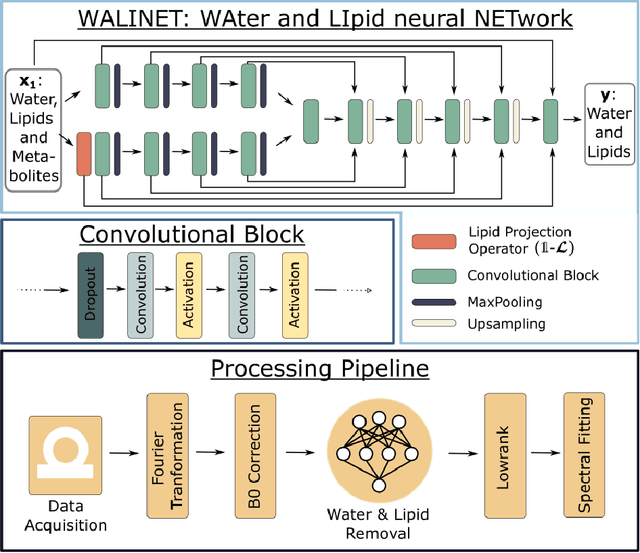

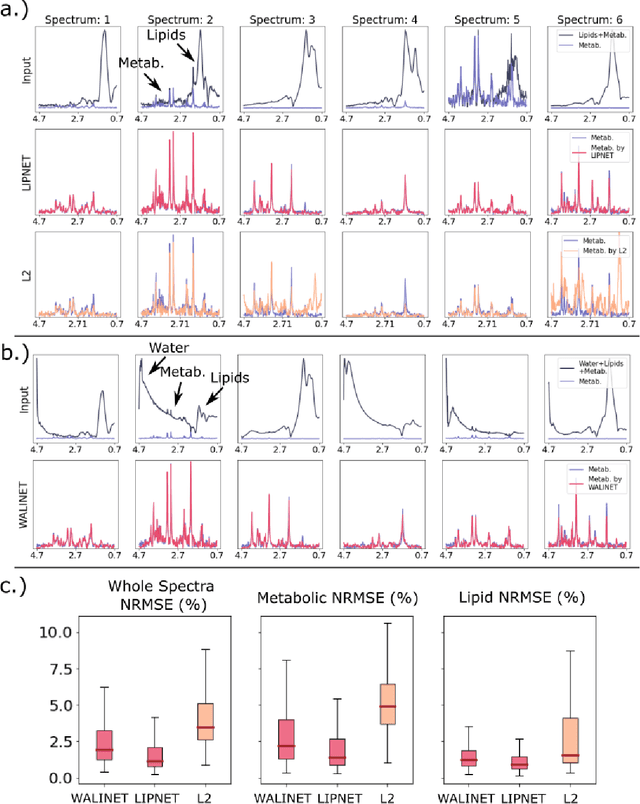
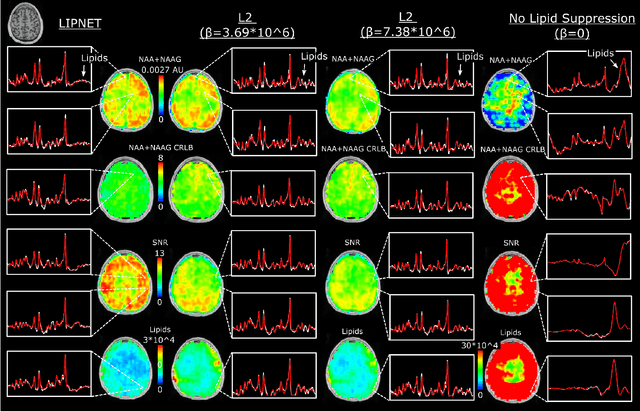
Abstract:Purpose. Proton Magnetic Resonance Spectroscopic Imaging (1H-MRSI) provides non-invasive spectral-spatial mapping of metabolism. However, long-standing problems in whole-brain 1H-MRSI are spectral overlap of metabolite peaks with large lipid signal from scalp, and overwhelming water signal that distorts spectra. Fast and effective methods are needed for high-resolution 1H-MRSI to accurately remove lipid and water signals while preserving the metabolite signal. The potential of supervised neural networks for this task remains unexplored, despite their success for other MRSI processing. Methods. We introduce a deep-learning method based on a modified Y-NET network for water and lipid removal in whole-brain 1H-MRSI. The WALINET (WAter and LIpid neural NETwork) was compared to conventional methods such as the state-of-the-art lipid L2 regularization and Hankel-Lanczos singular value decomposition (HLSVD) water suppression. Methods were evaluated on simulated and in-vivo whole-brain MRSI using NMRSE, SNR, CRLB, and FWHM metrics. Results. WALINET is significantly faster and needs 8s for high-resolution whole-brain MRSI, compared to 42 minutes for conventional HLSVD+L2. Quantitative analysis shows WALINET has better performance than HLSVD+L2: 1) more lipid removal with 41% lower NRMSE, 2) better metabolite signal preservation with 71% lower NRMSE in simulated data, 155% higher SNR and 50% lower CRLB in in-vivo data. Metabolic maps obtained by WALINET in healthy subjects and patients show better gray/white-matter contrast with more visible structural details. Conclusions. WALINET has superior performance for nuisance signal removal and metabolite quantification on whole-brain 1H-MRSI compared to conventional state-of-the-art techniques. This represents a new application of deep-learning for MRSI processing, with potential for automated high-throughput workflow.
Deep-ER: Deep Learning ECCENTRIC Reconstruction for fast high-resolution neurometabolic imaging
Sep 26, 2024
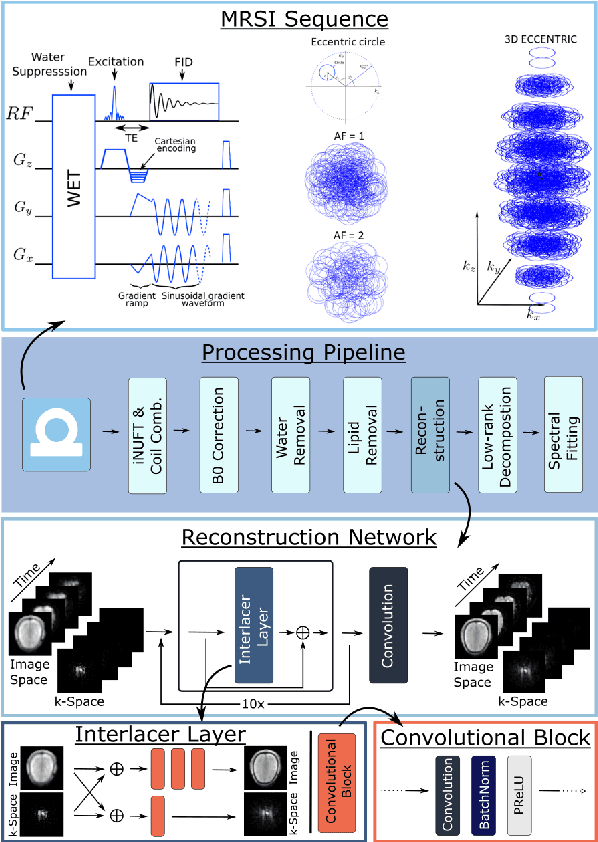
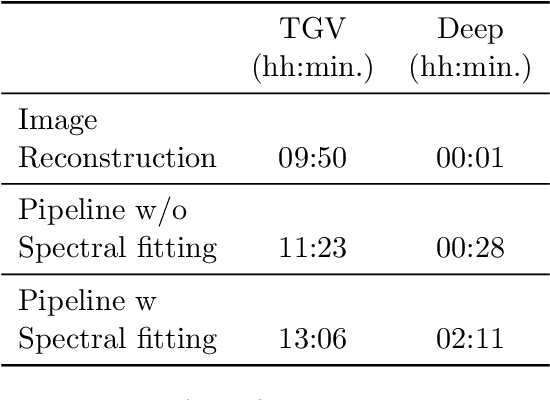

Abstract:Introduction: Altered neurometabolism is an important pathological mechanism in many neurological diseases and brain cancer, which can be mapped non-invasively by Magnetic Resonance Spectroscopic Imaging (MRSI). Advanced MRSI using non-cartesian compressed-sense acquisition enables fast high-resolution metabolic imaging but has lengthy reconstruction times that limits throughput and needs expert user interaction. Here, we present a robust and efficient Deep Learning reconstruction to obtain high-quality metabolic maps. Methods: Fast high-resolution whole-brain metabolic imaging was performed at 3.4 mm$^3$ isotropic resolution with acquisition times between 4:11-9:21 min:s using ECCENTRIC pulse sequence on a 7T MRI scanner. Data were acquired in a high-resolution phantom and 27 human participants, including 22 healthy volunteers and 5 glioma patients. A deep neural network using recurring interlaced convolutional layers with joint dual-space feature representation was developed for deep learning ECCENTRIC reconstruction (Deep-ER). 21 subjects were used for training and 6 subjects for testing. Deep-ER performance was compared to conventional iterative Total Generalized Variation reconstruction using image and spectral quality metrics. Results: Deep-ER demonstrated 600-fold faster reconstruction than conventional methods, providing improved spatial-spectral quality and metabolite quantification with 12%-45% (P<0.05) higher signal-to-noise and 8%-50% (P<0.05) smaller Cramer-Rao lower bounds. Metabolic images clearly visualize glioma tumor heterogeneity and boundary. Conclusion: Deep-ER provides efficient and robust reconstruction for sparse-sampled MRSI. The accelerated acquisition-reconstruction MRSI is compatible with high-throughput imaging workflow. It is expected that such improved performance will facilitate basic and clinical MRSI applications.
Registration by Regression (RbR): a framework for interpretable and flexible atlas registration
Apr 25, 2024


Abstract:In human neuroimaging studies, atlas registration enables mapping MRI scans to a common coordinate frame, which is necessary to aggregate data from multiple subjects. Machine learning registration methods have achieved excellent speed and accuracy but lack interpretability. More recently, keypoint-based methods have been proposed to tackle this issue, but their accuracy is still subpar, particularly when fitting nonlinear transforms. Here we propose Registration by Regression (RbR), a novel atlas registration framework that is highly robust and flexible, conceptually simple, and can be trained with cheaply obtained data. RbR predicts the (x,y,z) atlas coordinates for every voxel of the input scan (i.e., every voxel is a keypoint), and then uses closed-form expressions to quickly fit transforms using a wide array of possible deformation models, including affine and nonlinear (e.g., Bspline, Demons, invertible diffeomorphic models, etc.). Robustness is provided by the large number of voxels informing the registration and can be further increased by robust estimators like RANSAC. Experiments on independent public datasets show that RbR yields more accurate registration than competing keypoint approaches, while providing full control of the deformation model.
Boosting Skull-Stripping Performance for Pediatric Brain Images
Feb 26, 2024



Abstract:Skull-stripping is the removal of background and non-brain anatomical features from brain images. While many skull-stripping tools exist, few target pediatric populations. With the emergence of multi-institutional pediatric data acquisition efforts to broaden the understanding of perinatal brain development, it is essential to develop robust and well-tested tools ready for the relevant data processing. However, the broad range of neuroanatomical variation in the developing brain, combined with additional challenges such as high motion levels, as well as shoulder and chest signal in the images, leaves many adult-specific tools ill-suited for pediatric skull-stripping. Building on an existing framework for robust and accurate skull-stripping, we propose developmental SynthStrip (d-SynthStrip), a skull-stripping model tailored to pediatric images. This framework exposes networks to highly variable images synthesized from label maps. Our model substantially outperforms pediatric baselines across scan types and age cohorts. In addition, the <1-minute runtime of our tool compares favorably to the fastest baselines. We distribute our model at https://w3id.org/synthstrip.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge