Alex Fedorov
Emory University
Longitudinal Progression Prediction of Alzheimer's Disease with Tabular Foundation Model
Aug 25, 2025Abstract:Alzheimer's disease is a progressive neurodegenerative disorder that remains challenging to predict due to its multifactorial etiology and the complexity of multimodal clinical data. Accurate forecasting of clinically relevant biomarkers, including diagnostic and quantitative measures, is essential for effective monitoring of disease progression. This work introduces L2C-TabPFN, a method that integrates a longitudinal-to-cross-sectional (L2C) transformation with a pre-trained Tabular Foundation Model (TabPFN) to predict Alzheimer's disease outcomes using the TADPOLE dataset. L2C-TabPFN converts sequential patient records into fixed-length feature vectors, enabling robust prediction of diagnosis, cognitive scores, and ventricular volume. Experimental results demonstrate that, while L2C-TabPFN achieves competitive performance on diagnostic and cognitive outcomes, it provides state-of-the-art results in ventricular volume prediction. This key imaging biomarker reflects neurodegeneration and progression in Alzheimer's disease. These findings highlight the potential of tabular foundational models for advancing longitudinal prediction of clinically relevant imaging markers in Alzheimer's disease.
Towards Synthesizing Normative Data for Cognitive Assessments Using Generative Multimodal Large Language Models
Aug 25, 2025Abstract:Cognitive assessments require normative data as essential benchmarks for evaluating individual performance. Hence, developing new cognitive tests based on novel image stimuli is challenging due to the lack of readily available normative data. Traditional data collection methods are costly, time-consuming, and infrequently updated, limiting their practical utility. Recent advancements in generative multimodal large language models (MLLMs) offer a new approach to generate synthetic normative data from existing cognitive test images. We investigated the feasibility of using MLLMs, specifically GPT-4o and GPT-4o-mini, to synthesize normative textual responses for established image-based cognitive assessments, such as the "Cookie Theft" picture description task. Two distinct prompting strategies-naive prompts with basic instructions and advanced prompts enriched with contextual guidance-were evaluated. Responses were analyzed using embeddings to assess their capacity to distinguish diagnostic groups and demographic variations. Performance metrics included BLEU, ROUGE, BERTScore, and an LLM-as-a-judge evaluation. Advanced prompting strategies produced synthetic responses that more effectively distinguished between diagnostic groups and captured demographic diversity compared to naive prompts. Superior models generated responses exhibiting higher realism and diversity. BERTScore emerged as the most reliable metric for contextual similarity assessment, while BLEU was less effective for evaluating creative outputs. The LLM-as-a-judge approach provided promising preliminary validation results. Our study demonstrates that generative multimodal LLMs, guided by refined prompting methods, can feasibly generate robust synthetic normative data for existing cognitive tests, thereby laying the groundwork for developing novel image-based cognitive assessments without the traditional limitations.
Learning ECG Representations via Poly-Window Contrastive Learning
Aug 21, 2025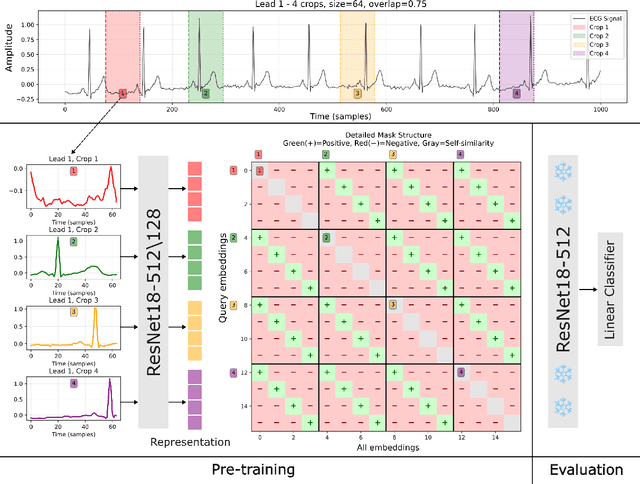
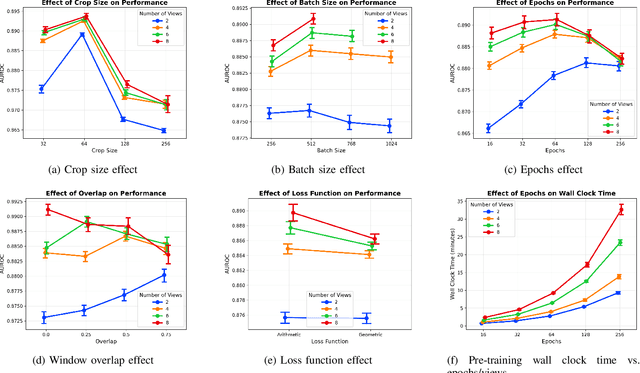
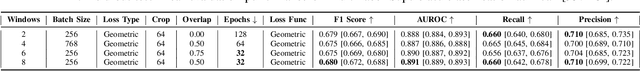

Abstract:Electrocardiogram (ECG) analysis is foundational for cardiovascular disease diagnosis, yet the performance of deep learning models is often constrained by limited access to annotated data. Self-supervised contrastive learning has emerged as a powerful approach for learning robust ECG representations from unlabeled signals. However, most existing methods generate only pairwise augmented views and fail to leverage the rich temporal structure of ECG recordings. In this work, we present a poly-window contrastive learning framework. We extract multiple temporal windows from each ECG instance to construct positive pairs and maximize their agreement via statistics. Inspired by the principle of slow feature analysis, our approach explicitly encourages the model to learn temporally invariant and physiologically meaningful features that persist across time. We validate our approach through extensive experiments and ablation studies on the PTB-XL dataset. Our results demonstrate that poly-window contrastive learning consistently outperforms conventional two-view methods in multi-label superclass classification, achieving higher AUROC (0.891 vs. 0.888) and F1 scores (0.680 vs. 0.679) while requiring up to four times fewer pre-training epochs (32 vs. 128) and 14.8% in total wall clock pre-training time reduction. Despite processing multiple windows per sample, we achieve a significant reduction in the number of training epochs and total computation time, making our method practical for training foundational models. Through extensive ablations, we identify optimal design choices and demonstrate robustness across various hyperparameters. These findings establish poly-window contrastive learning as a highly efficient and scalable paradigm for automated ECG analysis and provide a promising general framework for self-supervised representation learning in biomedical time-series data.
MindGrab for BrainChop: Fast and Accurate Skull Stripping for Command Line and Browser
Jun 13, 2025Abstract:We developed MindGrab, a parameter- and memory-efficient deep fully-convolutional model for volumetric skull-stripping in head images of any modality. Its architecture, informed by a spectral interpretation of dilated convolutions, was trained exclusively on modality-agnostic synthetic data. MindGrab was evaluated on a retrospective dataset of 606 multimodal adult-brain scans (T1, T2, DWI, MRA, PDw MRI, EPI, CT, PET) sourced from the SynthStrip dataset. Performance was benchmarked against SynthStrip, ROBEX, and BET using Dice scores, with Wilcoxon signed-rank significance tests. MindGrab achieved a mean Dice score of 95.9 with standard deviation (SD) 1.6 across modalities, significantly outperforming classical methods (ROBEX: 89.1 SD 7.7, P < 0.05; BET: 85.2 SD 14.4, P < 0.05). Compared to SynthStrip (96.5 SD 1.1, P=0.0352), MindGrab delivered equivalent or superior performance in nearly half of the tested scenarios, with minor differences (<3% Dice) in the others. MindGrab utilized 95% fewer parameters (146,237 vs. 2,566,561) than SynthStrip. This efficiency yielded at least 2x faster inference, 50% lower memory usage on GPUs, and enabled exceptional performance (e.g., 10-30x speedup, and up to 30x memory reduction) and accessibility on a wider range of hardware, including systems without high-end GPUs. MindGrab delivers state-of-the-art accuracy with dramatically lower resource demands, supported in brainchop-cli (https://pypi.org/project/brainchop/) and at brainchop.org.
State-of-the-Art Stroke Lesion Segmentation at 1/1000th of Parameters
Mar 07, 2025



Abstract:Efficient and accurate whole-brain lesion segmentation remains a challenge in medical image analysis. In this work, we revisit MeshNet, a parameter-efficient segmentation model, and introduce a novel multi-scale dilation pattern with an encoder-decoder structure. This innovation enables capturing broad contextual information and fine-grained details without traditional downsampling, upsampling, or skip-connections. Unlike previous approaches processing subvolumes or slices, we operate directly on whole-brain $256^3$ MRI volumes. Evaluations on the Aphasia Recovery Cohort (ARC) dataset demonstrate that MeshNet achieves superior or comparable DICE scores to state-of-the-art architectures such as MedNeXt and U-MAMBA at 1/1000th of parameters. Our results validate MeshNet's strong balance of efficiency and performance, making it particularly suitable for resource-limited environments such as web-based applications and opening new possibilities for the widespread deployment of advanced medical image analysis tools.
Early Risk Prediction of Pediatric Cardiac Arrest from Electronic Health Records via Multimodal Fused Transformer
Feb 11, 2025Abstract:Early prediction of pediatric cardiac arrest (CA) is critical for timely intervention in high-risk intensive care settings. We introduce PedCA-FT, a novel transformer-based framework that fuses tabular view of EHR with the derived textual view of EHR to fully unleash the interactions of high-dimensional risk factors and their dynamics. By employing dedicated transformer modules for each modality view, PedCA-FT captures complex temporal and contextual patterns to produce robust CA risk estimates. Evaluated on a curated pediatric cohort from the CHOA-CICU database, our approach outperforms ten other artificial intelligence models across five key performance metrics and identifies clinically meaningful risk factors. These findings underscore the potential of multimodal fusion techniques to enhance early CA detection and improve patient care.
Self-supervised multimodal neuroimaging yields predictive representations for a spectrum of Alzheimer's phenotypes
Sep 07, 2022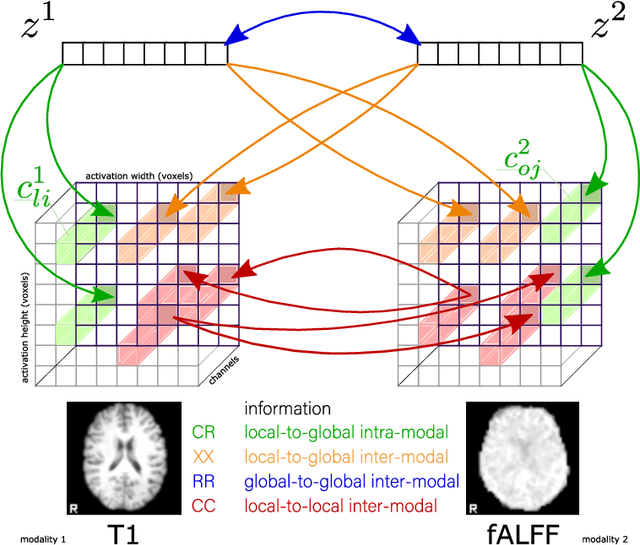
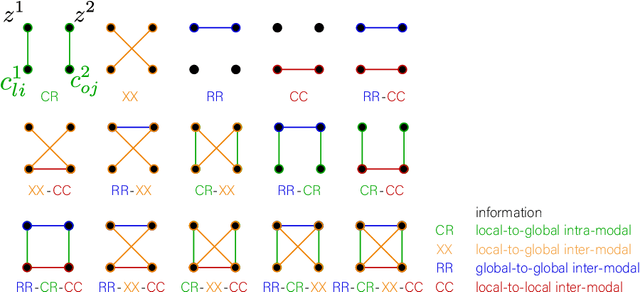
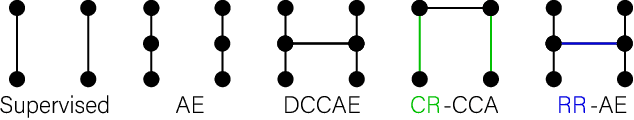
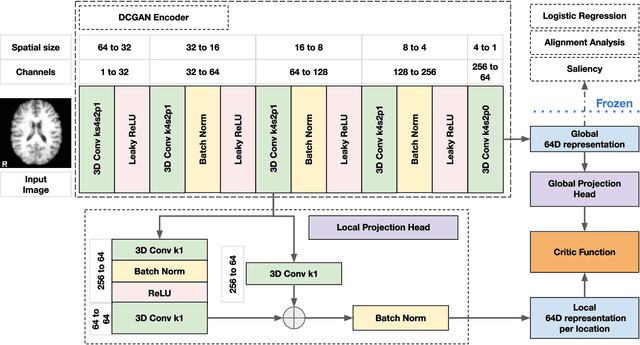
Abstract:Recent neuroimaging studies that focus on predicting brain disorders via modern machine learning approaches commonly include a single modality and rely on supervised over-parameterized models.However, a single modality provides only a limited view of the highly complex brain. Critically, supervised models in clinical settings lack accurate diagnostic labels for training. Coarse labels do not capture the long-tailed spectrum of brain disorder phenotypes, which leads to a loss of generalizability of the model that makes them less useful in diagnostic settings. This work presents a novel multi-scale coordinated framework for learning multiple representations from multimodal neuroimaging data. We propose a general taxonomy of informative inductive biases to capture unique and joint information in multimodal self-supervised fusion. The taxonomy forms a family of decoder-free models with reduced computational complexity and a propensity to capture multi-scale relationships between local and global representations of the multimodal inputs. We conduct a comprehensive evaluation of the taxonomy using functional and structural magnetic resonance imaging (MRI) data across a spectrum of Alzheimer's disease phenotypes and show that self-supervised models reveal disorder-relevant brain regions and multimodal links without access to the labels during pre-training. The proposed multimodal self-supervised learning yields representations with improved classification performance for both modalities. The concomitant rich and flexible unsupervised deep learning framework captures complex multimodal relationships and provides predictive performance that meets or exceeds that of a more narrow supervised classification analysis. We present elaborate quantitative evidence of how this framework can significantly advance our search for missing links in complex brain disorders.
Pipeline-Invariant Representation Learning for Neuroimaging
Aug 27, 2022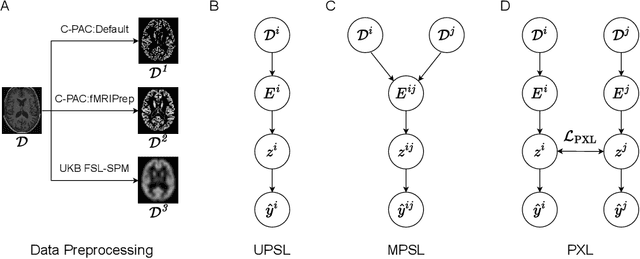

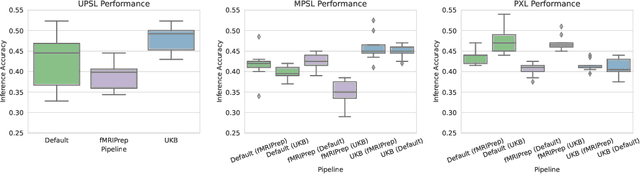
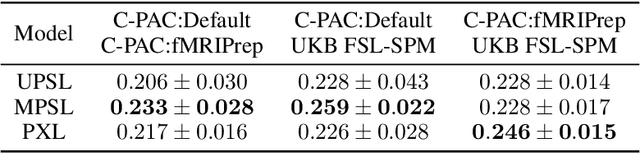
Abstract:Deep learning has been widely applied in neuroimaging, including to predicting brain-phenotype relationships from magnetic resonance imaging (MRI) volumes. MRI data usually requires extensive preprocessing before it is ready for modeling, even via deep learning, in part due to its high dimensionality and heterogeneity. A growing array of MRI preprocessing pipelines have been developed each with its own strengths and limitations. Recent studies have shown that pipeline-related variation may lead to different scientific findings, even when using the identical data. Meanwhile, the machine learning community has emphasized the importance of shifting from model-centric to data-centric approaches given that data quality plays an essential role in deep learning applications. Motivated by this idea, we first evaluate how preprocessing pipeline selection can impact the downstream performance of a supervised learning model. We next propose two pipeline-invariant representation learning methodologies, MPSL and PXL, to improve consistency in classification performance and to capture similar neural network representations between pipeline pairs. Using 2000 human subjects from the UK Biobank dataset, we demonstrate that both models present unique advantages, in particular that MPSL can be used to improve out-of-sample generalization to new pipelines, while PXL can be used to improve predictive performance consistency and representational similarity within a closed pipeline set. These results suggest that our proposed models can be applied to overcome pipeline-related biases and to improve reproducibility in neuroimaging prediction tasks.
Tasting the cake: evaluating self-supervised generalization on out-of-distribution multimodal MRI data
Apr 20, 2021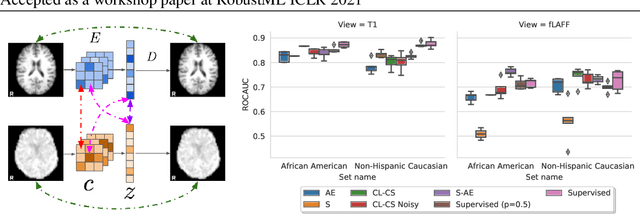
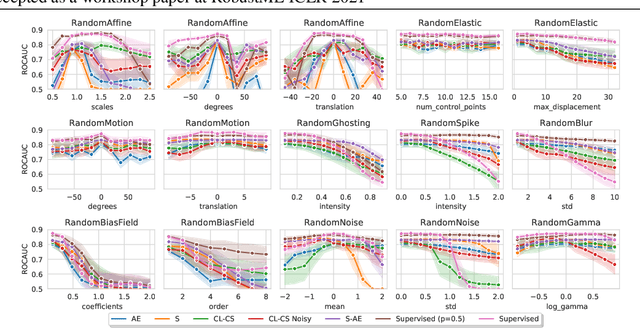
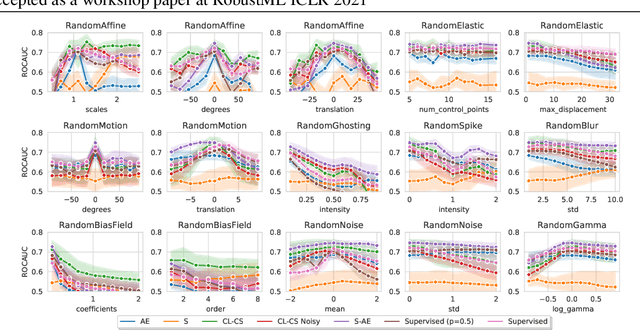

Abstract:Self-supervised learning has enabled significant improvements on natural image benchmarks. However, there is less work in the medical imaging domain in this area. The optimal models have not yet been determined among the various options. Moreover, little work has evaluated the current applicability limits of novel self-supervised methods. In this paper, we evaluate a range of current contrastive self-supervised methods on out-of-distribution generalization in order to evaluate their applicability to medical imaging. We show that self-supervised models are not as robust as expected based on their results in natural imaging benchmarks and can be outperformed by supervised learning with dropout. We also show that this behavior can be countered with extensive augmentation. Our results highlight the need for out-of-distribution generalization standards and benchmarks to adopt the self-supervised methods in the medical imaging community.
Taxonomy of multimodal self-supervised representation learning
Dec 29, 2020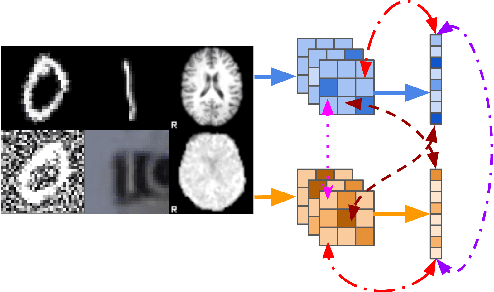
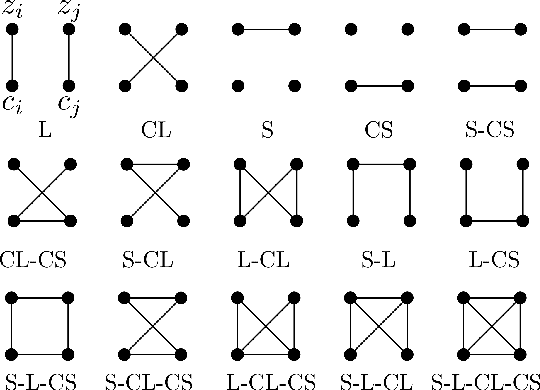
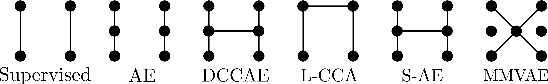
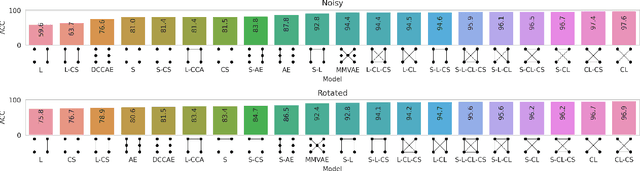
Abstract:Sensory input from multiple sources is crucial for robust and coherent human perception. Different sources contribute complementary explanatory factors and get combined based on factors they share. This system motivated the design of powerful unsupervised representation-learning algorithms. In this paper, we unify recent work on multimodal self-supervised learning under a single framework. Observing that most self-supervised methods optimize similarity metrics between a set of model components, we propose a taxonomy of all reasonable ways to organize this process. We empirically show on two versions of multimodal MNIST and a multimodal brain imaging dataset that (1) multimodal contrastive learning has significant benefits over its unimodal counterpart, (2) the specific composition of multiple contrastive objectives is critical to performance on a downstream task, (3) maximization of the similarity between representations has a regularizing effect on a neural network, which sometimes can lead to reduced downstream performance but still can reveal multimodal relations. Consequently, we outperform previous unsupervised encoder-decoder methods based on CCA or variational mixtures MMVAE on various datasets on linear evaluation protocol.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge