Vince Calhoun
Breast Cancer Detection in Thermographic Images via Diffusion-Based Augmentation and Nonlinear Feature Fusion
Sep 08, 2025Abstract:Data scarcity hinders deep learning for medical imaging. We propose a framework for breast cancer classification in thermograms that addresses this using a Diffusion Probabilistic Model (DPM) for data augmentation. Our DPM-based augmentation is shown to be superior to both traditional methods and a ProGAN baseline. The framework fuses deep features from a pre-trained ResNet-50 with handcrafted nonlinear features (e.g., Fractal Dimension) derived from U-Net segmented tumors. An XGBoost classifier trained on these fused features achieves 98.0\% accuracy and 98.1\% sensitivity. Ablation studies and statistical tests confirm that both the DPM augmentation and the nonlinear feature fusion are critical, statistically significant components of this success. This work validates the synergy between advanced generative models and interpretable features for creating highly accurate medical diagnostic tools.
Causal Graph Recovery in Neuroimaging through Answer Set Programming
Jun 10, 2025Abstract:Learning graphical causal structures from time series data presents significant challenges, especially when the measurement frequency does not match the causal timescale of the system. This often leads to a set of equally possible underlying causal graphs due to information loss from sub-sampling (i.e., not observing all possible states of the system throughout time). Our research addresses this challenge by incorporating the effects of sub-sampling in the derivation of causal graphs, resulting in more accurate and intuitive outcomes. We use a constraint optimization approach, specifically answer set programming (ASP), to find the optimal set of answers. ASP not only identifies the most probable underlying graph, but also provides an equivalence class of possible graphs for expert selection. In addition, using ASP allows us to leverage graph theory to further prune the set of possible solutions, yielding a smaller, more accurate answer set significantly faster than traditional approaches. We validate our approach on both simulated data and empirical structural brain connectivity, and demonstrate its superiority over established methods in these experiments. We further show how our method can be used as a meta-approach on top of established methods to obtain, on average, 12% improvement in F1 score. In addition, we achieved state of the art results in terms of precision and recall of reconstructing causal graph from sub-sampled time series data. Finally, our method shows robustness to varying degrees of sub-sampling on realistic simulations, whereas other methods perform worse for higher rates of sub-sampling.
Mapping minds not averages: a scalable subject-specific manifold learning framework for neuroimaging data
Apr 30, 2025Abstract:Mental and cognitive representations are believed to reside on low-dimensional, non-linear manifolds embedded within high-dimensional brain activity. Uncovering these manifolds is key to understanding individual differences in brain function, yet most existing machine learning methods either rely on population-level spatial alignment or assume data that is temporally structured, either because data is aligned among subjects or because event timings are known. We introduce a manifold learning framework that can capture subject-specific spatial variations across both structured and temporally unstructured neuroimaging data. On simulated data and two naturalistic fMRI datasets (Sherlock and Forrest Gump), our framework outperforms group-based baselines by recovering more accurate and individualized representations. We further show that the framework scales efficiently to large datasets and generalizes well to new subjects. To test this, we apply the framework to temporally unstructured resting-state fMRI data from individuals with schizophrenia and healthy controls. We further apply our method to a large resting-state fMRI dataset comprising individuals with schizophrenia and controls. In this setting, we demonstrate that the framework scales efficiently to large populations and generalizes robustly to unseen subjects. The learned subject-specific spatial maps our model finds reveal clinically relevant patterns, including increased activation in the basal ganglia, visual, auditory, and somatosensory regions, and decreased activation in the insula, inferior frontal gyrus, and angular gyrus. These findings suggest that our framework can uncover clinically relevant subject-specific brain activity patterns. Our approach thus provides a scalable and individualized framework for modeling brain activity, with applications in computational neuroscience and clinical research.
Rethinking Functional Brain Connectome Analysis: Do Graph Deep Learning Models Help?
Jan 28, 2025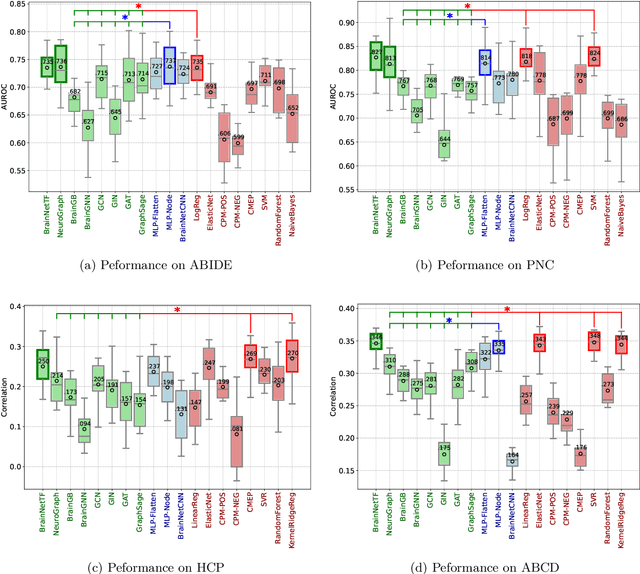
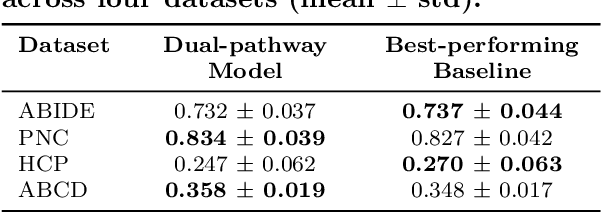
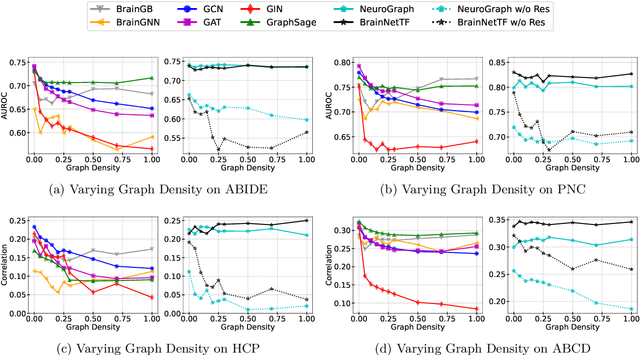
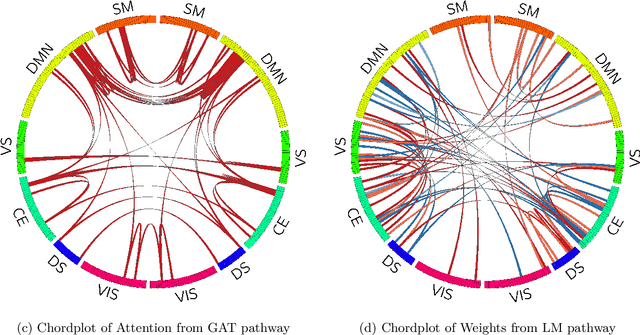
Abstract:Functional brain connectome is crucial for deciphering the neural mechanisms underlying cognitive functions and neurological disorders. Graph deep learning models have recently gained tremendous popularity in this field. However, their actual effectiveness in modeling the brain connectome remains unclear. In this study, we re-examine graph deep learning models based on four large-scale neuroimaging studies encompassing diverse cognitive and clinical outcomes. Surprisingly, we find that the message aggregation mechanism, a hallmark of graph deep learning models, does not help with predictive performance as typically assumed, but rather consistently degrades it. To address this issue, we propose a hybrid model combining a linear model with a graph attention network through dual pathways, achieving robust predictions and enhanced interpretability by revealing both localized and global neural connectivity patterns. Our findings urge caution in adopting complex deep learning models for functional brain connectome analysis, emphasizing the need for rigorous experimental designs to establish tangible performance gains and perhaps more importantly, to pursue improvements in model interpretability.
Hierarchical Spatio-Temporal State-Space Modeling for fMRI Analysis
Aug 23, 2024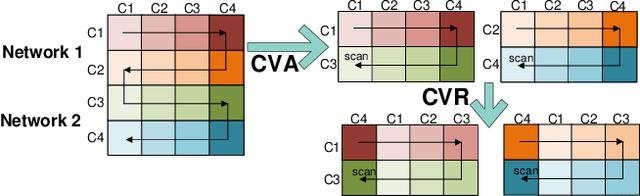
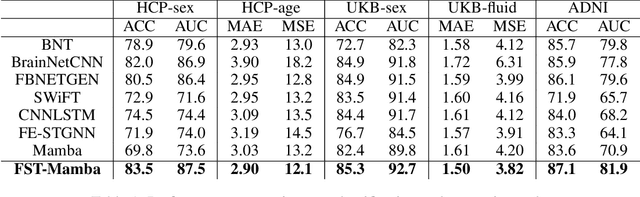
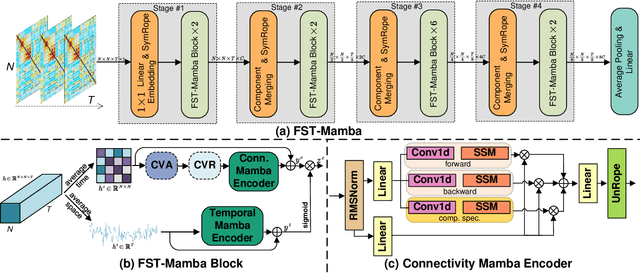

Abstract:Recent advances in deep learning structured state space models, especially the Mamba architecture, have demonstrated remarkable performance improvements while maintaining linear complexity. In this study, we introduce functional spatiotemporal Mamba (FST-Mamba), a Mamba-based model designed for discovering neurological biomarkers using functional magnetic resonance imaging (fMRI). We focus on dynamic functional network connectivity (dFNC) derived from fMRI and propose a hierarchical spatiotemporal Mamba-based network that processes spatial and temporal information separately using Mamba-based encoders. Leveraging the topological uniqueness of the FNC matrix, we introduce a component-wise varied-scale aggregation (CVA) mechanism to aggregate connectivity across individual components within brain networks, enabling the model to capture both inter-component and inter-network information. To better handle the FNC data, we develop a new component-specific scanning order. Additionally, we propose symmetric rotary position encoding (SymRope) to encode the relative positions of each functional connection while considering the symmetric nature of the FNC matrix. Experimental results demonstrate significant improvements in the proposed FST-Mamba model on various brain-based classification and regression tasks. Our work reveals the substantial potential of attention-free sequence modeling in brain discovery.
Spatial Sequence Attention Network for Schizophrenia Classification from Structural Brain MR Images
Jun 18, 2024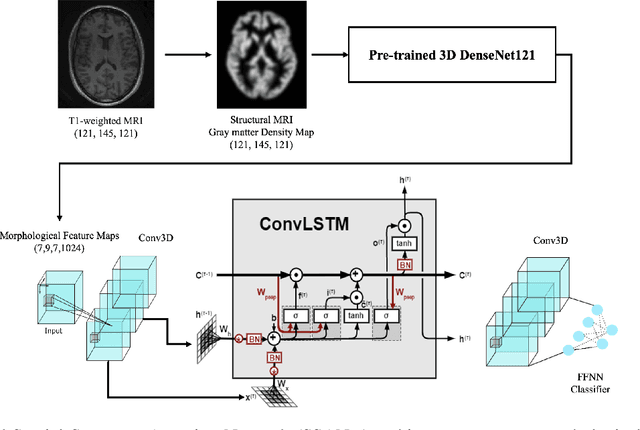
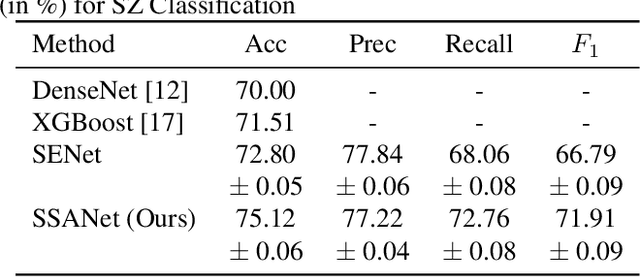
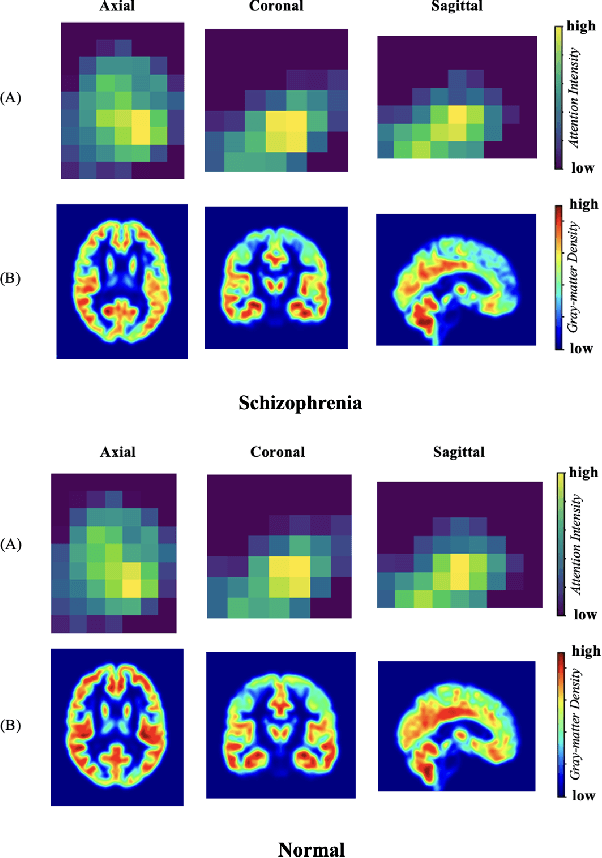
Abstract:Schizophrenia is a debilitating, chronic mental disorder that significantly impacts an individual's cognitive abilities, behavior, and social interactions. It is characterized by subtle morphological changes in the brain, particularly in the gray matter. These changes are often imperceptible through manual observation, demanding an automated approach to diagnosis. This study introduces a deep learning methodology for the classification of individuals with Schizophrenia. We achieve this by implementing a diversified attention mechanism known as Spatial Sequence Attention (SSA) which is designed to extract and emphasize significant feature representations from structural MRI (sMRI). Initially, we employ the transfer learning paradigm by leveraging pre-trained DenseNet to extract initial feature maps from the final convolutional block which contains morphological alterations associated with Schizophrenia. These features are further processed by the proposed SSA to capture and emphasize intricate spatial interactions and relationships across volumes within the brain. Our experimental studies conducted on a clinical dataset have revealed that the proposed attention mechanism outperforms the existing Squeeze & Excitation Network for Schizophrenia classification.
Unmasking Efficiency: Learning Salient Sparse Models in Non-IID Federated Learning
May 15, 2024



Abstract:In this work, we propose Salient Sparse Federated Learning (SSFL), a streamlined approach for sparse federated learning with efficient communication. SSFL identifies a sparse subnetwork prior to training, leveraging parameter saliency scores computed separately on local client data in non-IID scenarios, and then aggregated, to determine a global mask. Only the sparse model weights are communicated each round between the clients and the server. We validate SSFL's effectiveness using standard non-IID benchmarks, noting marked improvements in the sparsity--accuracy trade-offs. Finally, we deploy our method in a real-world federated learning framework and report improvement in communication time.
Brain Networks and Intelligence: A Graph Neural Network Based Approach to Resting State fMRI Data
Nov 06, 2023Abstract:Resting-state functional magnetic resonance imaging (rsfMRI) is a powerful tool for investigating the relationship between brain function and cognitive processes as it allows for the functional organization of the brain to be captured without relying on a specific task or stimuli. In this paper, we present a novel modeling architecture called BrainRGIN for predicting intelligence (fluid, crystallized, and total intelligence) using graph neural networks on rsfMRI derived static functional network connectivity matrices. Extending from the existing graph convolution networks, our approach incorporates a clustering-based embedding and graph isomorphism network in the graph convolutional layer to reflect the nature of the brain sub-network organization and efficient network expression, in combination with TopK pooling and attention-based readout functions. We evaluated our proposed architecture on a large dataset, specifically the Adolescent Brain Cognitive Development Dataset, and demonstrated its effectiveness in predicting individual differences in intelligence. Our model achieved lower mean squared errors and higher correlation scores than existing relevant graph architectures and other traditional machine learning models for all of the intelligence prediction tasks. The middle frontal gyrus exhibited a significant contribution to both fluid and crystallized intelligence, suggesting their pivotal role in these cognitive processes. Total composite scores identified a diverse set of brain regions to be relevant which underscores the complex nature of total intelligence.
Cross-Modal Synthesis of Structural MRI and Functional Connectivity Networks via Conditional ViT-GANs
Sep 15, 2023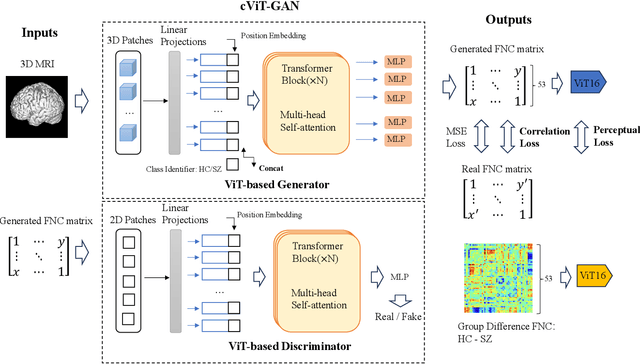
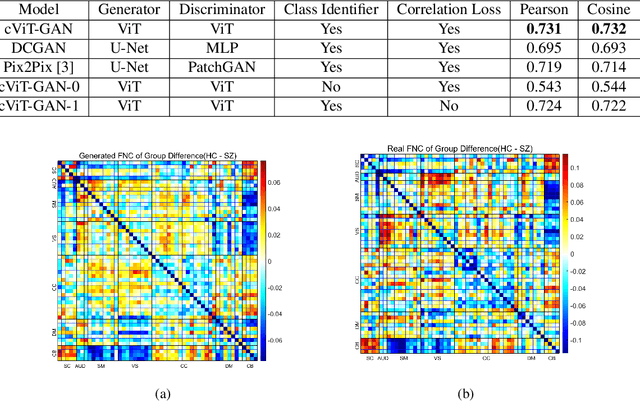
Abstract:The cross-modal synthesis between structural magnetic resonance imaging (sMRI) and functional network connectivity (FNC) is a relatively unexplored area in medical imaging, especially with respect to schizophrenia. This study employs conditional Vision Transformer Generative Adversarial Networks (cViT-GANs) to generate FNC data based on sMRI inputs. After training on a comprehensive dataset that included both individuals with schizophrenia and healthy control subjects, our cViT-GAN model effectively synthesized the FNC matrix for each subject, and then formed a group difference FNC matrix, obtaining a Pearson correlation of 0.73 with the actual FNC matrix. In addition, our FNC visualization results demonstrate significant correlations in particular subcortical brain regions, highlighting the model's capability of capturing detailed structural-functional associations. This performance distinguishes our model from conditional CNN-based GAN alternatives such as Pix2Pix. Our research is one of the first attempts to link sMRI and FNC synthesis, setting it apart from other cross-modal studies that concentrate on T1- and T2-weighted MR images or the fusion of MRI and CT scans.
Learning low-dimensional dynamics from whole-brain data improves task capture
May 18, 2023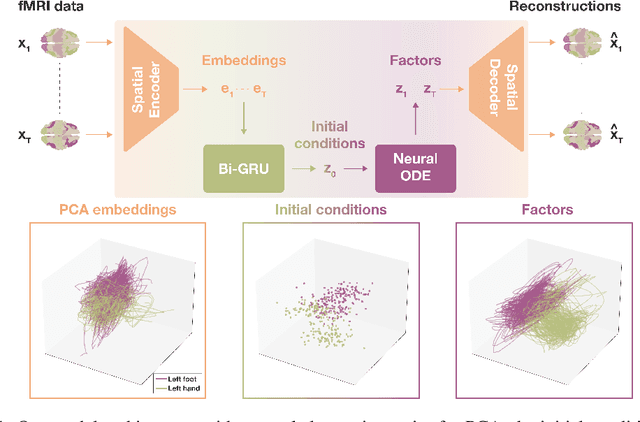
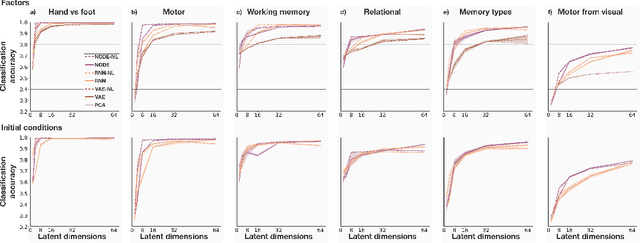
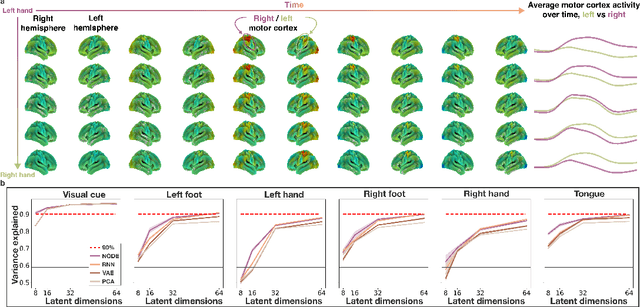
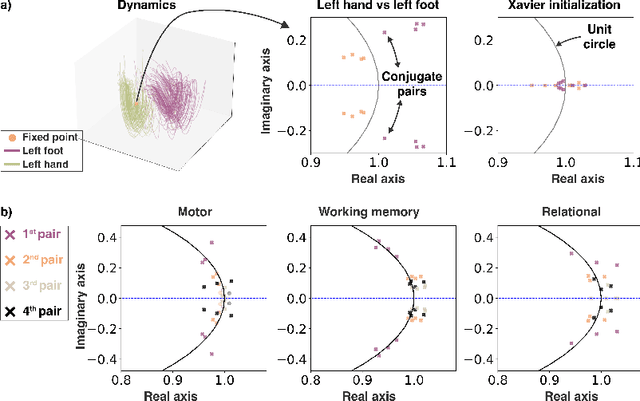
Abstract:The neural dynamics underlying brain activity are critical to understanding cognitive processes and mental disorders. However, current voxel-based whole-brain dimensionality reduction techniques fall short of capturing these dynamics, producing latent timeseries that inadequately relate to behavioral tasks. To address this issue, we introduce a novel approach to learning low-dimensional approximations of neural dynamics by using a sequential variational autoencoder (SVAE) that represents the latent dynamical system via a neural ordinary differential equation (NODE). Importantly, our method finds smooth dynamics that can predict cognitive processes with accuracy higher than classical methods. Our method also shows improved spatial localization to task-relevant brain regions and identifies well-known structures such as the motor homunculus from fMRI motor task recordings. We also find that non-linear projections to the latent space enhance performance for specific tasks, offering a promising direction for future research. We evaluate our approach on various task-fMRI datasets, including motor, working memory, and relational processing tasks, and demonstrate that it outperforms widely used dimensionality reduction techniques in how well the latent timeseries relates to behavioral sub-tasks, such as left-hand or right-hand tapping. Additionally, we replace the NODE with a recurrent neural network (RNN) and compare the two approaches to understand the importance of explicitly learning a dynamical system. Lastly, we analyze the robustness of the learned dynamical systems themselves and find that their fixed points are robust across seeds, highlighting our method's potential for the analysis of cognitive processes as dynamical systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge